 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Zmw_sc01257.1.g00110.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; PACMAD clade; Chloridoideae; Zoysieae; Zoysiinae; Zoysia
|
||||||||
| Family | EIL | ||||||||
| Protein Properties | Length: 612aa MW: 68484.9 Da PI: 5.731 | ||||||||
| Description | EIL family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | EIN3 | 414.6 | 1.1e-126 | 38 | 416 | 1 | 351 |
XXXXXXXXXXXXXXXXXXXXXXX..XXXXX.XXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXX CS
EIN3 1 eelkkrmwkdqmllkrlkerkkqlledkeaatgakksnksneqarrkkmsraQDgiLkYMlkemevcnaqGfvYgiipe 79
eel krmwkd+++l+r+ker+++l ++a+++++k + ++qa rkkmsr DgiL+YMlk+mevcna+GfvYgiip+
Zmw_sc01257.1.g00110.1.es.mkhc 38 EELAKRMWKDRVRLRRIKERQQRLA-LQKADLEKAKLKPVSDQAMRKKMSRVHDGILRYMLKMMEVCNARGFVYGIIPD 115
8*********************976.55556************************************************ PP
XXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXX....XX----STTS-HHHHHHHHHHHSSSSSS- CS
EIN3 80 kgkpvegasdsLraWWkekvefdrngpaaiskyqaknlilsgesslqtersseshslselqDTtlgSLLsalmqhcdpp 158
kgkpv+gasd++raWWkekv+fd+ngpaai+ky+++n+++++ +s + ++ hsl++lqD+tlgSLLs+lmqhc+pp
Zmw_sc01257.1.g00110.1.es.mkhc 116 KGKPVSGASDNIRAWWKEKVKFDKNGPAAIAKYESENMAVADPQSG---GVKNLHSLMDLQDATLGSLLSSLMQHCNPP 191
***************************************9987777...89**************************** PP
TTS--TTT--HHHH---S--HHHHHHT--TT--.-----GGG--HHHHHHHHHHHHHHTGGGHHHHHHTTTTSSSSTTT CS
EIN3 159 qrrfplekgvepPWWPtGkelwwgelglskdqgtppykkphdlkkawkvsvLtavikhmsptieeirelerqskylqdk 237
qr++plekg++pPWWP+G+e+ww +gl++ q ppykkphdlkk+wkv+vLt vikhmsp++++ir++ r+sk+lqdk
Zmw_sc01257.1.g00110.1.es.mkhc 192 QRKYPLEKGTPPPWWPSGNEEWWIDMGLPTGQV-PPYKKPHDLKKVWKVGVLTGVIKHMSPNFDKIRNHIRKSKCLQDK 269
******************************999.9******************************************** PP
--SHHHHHHHHHHTTTTT-S--XXXX.XX....XXXXXXXXXXXXXXXXXXXXXXXXXX.XXXXXXXXXX......... CS
EIN3 238 msakesfallsvlnqeekecatvsah.ss....slrkqspkvtlsceqkedvegkkeskikhvqavktta......... 302
m+akes ++l vl++ee++++ s+ s+ +++ t+s+++++d +g ++ ++++++++ +
Zmw_sc01257.1.g00110.1.es.mkhc 270 MTAKESLIWLGVLRREEQLVHGPDNGvSEithhSVAEGRYGDTHSNSDEYDIDGFEDAPLSTSSKDDEPDpspiaqspe 348
******************9987776653366678888999*************87777779888888888888888888 PP
........................XXXXXXXXXXXXXXXXXXXXX......XXXXXXX.XXXXXXXXXXXXXX CS
EIN3 303 ........................gfpvvrkrkkkpsesakvsskevsrtcqssqfrgsetelifadknsisq 351
g++ +k k+ +++ + ++s q+ ++++++++d+n+++q
Zmw_sc01257.1.g00110.1.es.mkhc 349 dnarkrgreraynkhpnqivlnkvGTKEPQKTKRARRRITVIES-----EAQRIDDAPESSRNMIPDMNQLDQ 416
88888888888888887776665444444444433333333322.....234444444567777777777666 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF04873 | 1.0E-123 | 38 | 286 | No hit | No description |
| Gene3D | G3DSA:1.10.3180.10 | 1.0E-68 | 162 | 290 | IPR023278 | Ethylene insensitive 3-like protein, DNA-binding domain |
| SuperFamily | SSF116768 | 1.57E-60 | 166 | 288 | IPR023278 | Ethylene insensitive 3-like protein, DNA-binding domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0042762 | Biological Process | regulation of sulfur metabolic process | ||||
| GO:0071281 | Biological Process | cellular response to iron ion | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 612 aa Download sequence Send to blast |
MEQLAMIASE LGDSSDLELE GIQNLNENDV SDEEIEPEEL AKRMWKDRVR LRRIKERQQR 60 LALQKADLEK AKLKPVSDQA MRKKMSRVHD GILRYMLKMM EVCNARGFVY GIIPDKGKPV 120 SGASDNIRAW WKEKVKFDKN GPAAIAKYES ENMAVADPQS GGVKNLHSLM DLQDATLGSL 180 LSSLMQHCNP PQRKYPLEKG TPPPWWPSGN EEWWIDMGLP TGQVPPYKKP HDLKKVWKVG 240 VLTGVIKHMS PNFDKIRNHI RKSKCLQDKM TAKESLIWLG VLRREEQLVH GPDNGVSEIT 300 HHSVAEGRYG DTHSNSDEYD IDGFEDAPLS TSSKDDEPDP SPIAQSPEDN ARKRGRERAY 360 NKHPNQIVLN KVGTKEPQKT KRARRRITVI ESEAQRIDDA PESSRNMIPD MNQLDQLETP 420 GLANQIVSFN HGGNMSDAFQ HRGDAQVQAH LPGAEVNSFG NAQAENATPV SIYMGGQHLP 480 YQNNDNTRSK SGNTFSVDAD SGLNNLPSSY QTLPPKQSLP LPMMDHHVVP MGTRAPADNG 540 PYNDQILTGG NSTSVPGDMQ QLMDYPFYGE QDKFVGSSFE GLPLDYISIS SPIPDIDDLL 600 LHDDDLMEYL GT |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1wij_A | 5e-66 | 169 | 295 | 12 | 140 | ETHYLENE-INSENSITIVE3-like 3 protein |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor that may be involved in the ethylene response pathway. {ECO:0000269|PubMed:9215635, ECO:0000269|PubMed:9851977}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00649 | PBM | Transfer from LOC_Os09g31400 | Download |
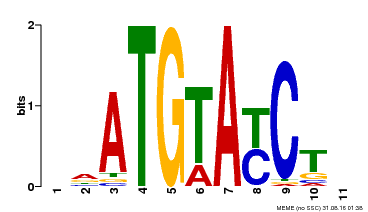
| |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_004957192.1 | 0.0 | ETHYLENE INSENSITIVE 3-like 3 protein | ||||
| Refseq | XP_004957193.1 | 0.0 | ETHYLENE INSENSITIVE 3-like 3 protein | ||||
| Swissprot | O23116 | 1e-140 | EIL3_ARATH; ETHYLENE INSENSITIVE 3-like 3 protein | ||||
| TrEMBL | A0A0A9DKK8 | 0.0 | A0A0A9DKK8_ARUDO; Uncharacterized protein | ||||
| STRING | Si029258m | 0.0 | (Setaria italica) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP3720 | 37 | 79 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



