 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Zmw_sc01982.1.g00170.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; PACMAD clade; Chloridoideae; Zoysieae; Zoysiinae; Zoysia
|
||||||||
| Family | bHLH | ||||||||
| Protein Properties | Length: 672aa MW: 72721.9 Da PI: 8.3269 | ||||||||
| Description | bHLH family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | HLH | 52.4 | 9.3e-17 | 287 | 333 | 4 | 55 |
HHHHHHHHHHHHHHHHHHHHHCTSCCC...TTS-STCHHHHHHHHHHHHHHH CS
HLH 4 ahnerErrRRdriNsafeeLrellPkaskapskKlsKaeiLekAveYIksLq 55
hn ErrRRdriN+++ L+el+P++ K +Ka++L +A+eY++sLq
Zmw_sc01982.1.g00170.1.am.mk 287 VHNLSERRRRDRINEKMKALQELIPHC-----NKTDKASMLDEAIEYLRSLQ 333
6*************************8.....6******************9 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:4.10.280.10 | 1.0E-20 | 280 | 340 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| CDD | cd00083 | 2.27E-18 | 280 | 337 | No hit | No description |
| SuperFamily | SSF47459 | 2.09E-20 | 281 | 343 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| PROSITE profile | PS50888 | 18.47 | 283 | 332 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Pfam | PF00010 | 3.6E-14 | 287 | 333 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SMART | SM00353 | 1.1E-18 | 289 | 338 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009704 | Biological Process | de-etiolation | ||||
| GO:0010161 | Biological Process | red light signaling pathway | ||||
| GO:0010244 | Biological Process | response to low fluence blue light stimulus by blue low-fluence system | ||||
| GO:0010600 | Biological Process | regulation of auxin biosynthetic process | ||||
| GO:0010928 | Biological Process | regulation of auxin mediated signaling pathway | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 672 aa Download sequence Send to blast |
MNQFVHDWAS SMGDAFAPLG FVPVIYGGLF LDDSCVIPFG SWLTSCESAM RFYSVEDDGL 60 MELLWCNGHV VMQSQAPRKP PRSDKEEAVQ GNEAEAWFQC PIEDPLERDL FSELFGETPA 120 DVPSRAACKE EEDCAAVTAP QLSRPNKACL EEIAGVSNCA VGKPDAGGDG EAATETGESS 180 MLTIGSSFCG SNHVQTTPRV VPLTPPRGGK AGARARDKRK QRDVAETEVR RTRAAGVFLP 240 RKRSITRPYR NNDTAAALQD VEFESAVVTC EPAQKLKTAK RRRAAEVHNL SERRRRDRIN 300 EKMKALQELI PHCNKTDKAS MLDEAIEYLR SLQLQLQVMW MGGGMATAAP VMFPAGVHQY 360 MQRMVGPPHV AAMPRLPPFM APPAVQSPPV GQMPVADPYA GCLAVDSLQT PGPMHYLQGV 420 NFYQRQSPAP PPPPVVPGGS LPAATALTLP SDNTLHKEHG MQCNSPATKQ NLDHFTRERS 480 LAIAIANHPF RQLVIRQLWS LRSFGCVELM DGAPYRRLEI RTPVPSASVP AAPFGTCWQT 540 RVPVSTLKKP GNGRSEQKGA AVTLSSHLCR PMSCRPGPRA PAPAAPSRAK TRARALPATT 600 SPWGMAMAAE RWTTAARVPA ADDDEAPGGT HCVGGGGDRC LSAVRNSSSS SSVRSITILA 660 LQISFQQQEK GQ |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 212 | 220 | ARARDKRKQ |
| 2 | 215 | 221 | RDKRKQR |
| 3 | 275 | 281 | LKTAKRR |
| 4 | 280 | 295 | RRRAAEVHNLSERRRR |
| 5 | 291 | 296 | ERRRRD |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00606 | ChIP-seq | Transfer from AT2G43010 | Download |
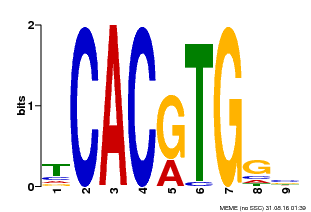
| |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_004963135.1 | 1e-131 | transcription factor PIF4 isoform X1 | ||||
| Refseq | XP_004963136.1 | 1e-131 | transcription factor PIF4 isoform X1 | ||||
| Refseq | XP_022680515.1 | 1e-131 | transcription factor PIF4 isoform X1 | ||||
| TrEMBL | K3Z6M7 | 1e-130 | K3Z6M7_SETIT; Uncharacterized protein | ||||
| STRING | Si022196m | 1e-131 | (Setaria italica) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP3107 | 36 | 77 |



