 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Zmw_sc02011.1.g00030.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; PACMAD clade; Chloridoideae; Zoysieae; Zoysiinae; Zoysia
|
||||||||
| Family | MYB_related | ||||||||
| Protein Properties | Length: 282aa MW: 30577.8 Da PI: 10.2919 | ||||||||
| Description | MYB_related family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 36.8 | 9.3e-12 | 53 | 104 | 1 | 47 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHT.....TTS-HHHHHHHHHHH CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmg.....kgRtlkqcksrwqky 47
+++WT+eE++ l ++v ++G g W+tI + R+ ++k++w+++
Zmw_sc02011.1.g00030.1.am.mkhc 53 KQKWTQEEEDALRRGVLKHGAGKWRTIQKDPEfsptlSSRSNIDLKDKWRNL 104
79***********************************9************96 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51294 | 10.905 | 48 | 108 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 2.23E-15 | 50 | 106 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 4.6E-8 | 52 | 107 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 1.5E-8 | 53 | 104 | IPR001005 | SANT/Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 3.5E-14 | 53 | 104 | IPR009057 | Homeodomain-like |
| CDD | cd11660 | 9.40E-20 | 54 | 104 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 282 aa Download sequence Send to blast |
ARERSRPPPG KDAHMNTRRV GEDRPLLVYV EGGWTLEKPQ IAGGEARAMG APKQKWTQEE 60 EDALRRGVLK HGAGKWRTIQ KDPEFSPTLS SRSNIDLKDK WRNLSFSAGQ GSREKTRVPK 120 ITGPSSSALP APQALLLRAS NKIAEASAPA DAEKKSQDGK ASPKFVDNKF YKVTDSFATK 180 TPALVKASTK KQKDPSKPLK ASKNLGIFAA VSPALEAAAA AAAKVADAEA KAHLAHEHMM 240 EADRILKMAE ETESLLTLAA EIYDRCSSGE IITVNPVSQK EF |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Binds preferentially double-stranded telomeric repeats, but may also bind to the single telomeric strand. {ECO:0000250}. | |||||
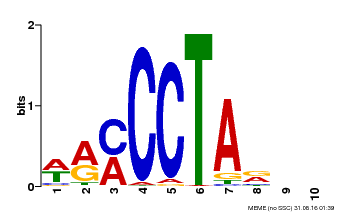
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00652 | PBM | Transfer from LOC_Os01g51154 | Download |
| |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | NP_001105225.1 | 2e-94 | single myb histone 3 | ||||
| Refseq | XP_025815372.1 | 8e-91 | glycylpeptide N-tetradecanoyltransferase 1 | ||||
| Swissprot | Q6WLH4 | 1e-95 | SMH3_MAIZE; Single myb histone 3 | ||||
| TrEMBL | A0A0A9PJS4 | 1e-104 | A0A0A9PJS4_ARUDO; Smh4 | ||||
| STRING | Pavir.Ea02570.1.p | 3e-87 | (Panicum virgatum) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP9247 | 34 | 42 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



