 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Zmw_sc02198.1.g00030.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; PACMAD clade; Chloridoideae; Zoysieae; Zoysiinae; Zoysia
|
||||||||
| Family | bHLH | ||||||||
| Protein Properties | Length: 523aa MW: 56644.2 Da PI: 7.1375 | ||||||||
| Description | bHLH family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | HLH | 52.9 | 6.7e-17 | 253 | 299 | 4 | 55 |
HHHHHHHHHHHHHHHHHHHHHCTSCCC...TTS-STCHHHHHHHHHHHHHHH CS
HLH 4 ahnerErrRRdriNsafeeLrellPkaskapskKlsKaeiLekAveYIksLq 55
hn ErrRRdriN+++ L+el+P++ K +Ka++L +A+eY++sLq
Zmw_sc02198.1.g00030.1.am.mk 253 VHNLSERRRRDRINEKMKALQELIPHC-----NKTDKASMLDEAIEYLRSLQ 299
6*************************8.....6******************9 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| CDD | cd00083 | 1.44E-18 | 246 | 303 | No hit | No description |
| Gene3D | G3DSA:4.10.280.10 | 7.2E-21 | 246 | 306 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SuperFamily | SSF47459 | 1.44E-20 | 247 | 309 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| PROSITE profile | PS50888 | 18.47 | 249 | 298 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Pfam | PF00010 | 2.6E-14 | 253 | 299 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SMART | SM00353 | 1.1E-18 | 255 | 304 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009704 | Biological Process | de-etiolation | ||||
| GO:0010161 | Biological Process | red light signaling pathway | ||||
| GO:0010244 | Biological Process | response to low fluence blue light stimulus by blue low-fluence system | ||||
| GO:0010600 | Biological Process | regulation of auxin biosynthetic process | ||||
| GO:0010928 | Biological Process | regulation of auxin mediated signaling pathway | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 523 aa Download sequence Send to blast |
MNQFVHDWAS SMGDAFAPLG VEDDGLMELL WCNGHVVMQS QAPRKPPRSD KEAAVQGNEA 60 EAWFQCPIED PLERDLFSEL FSETPADVPS RAACKEEEDC AAVTAPQLSR PNKACLEEIA 120 SVSNCAVGKP DAGGDGAAAT ETGESSMLTI GSSFCGSNHV QTTPRVVPPT PPRGEKAGAR 180 ARDKRKQRDV AETEVRRTRA AGAFLPRKRS ITRPYRNNDT AAALQDVEFE SAVVTCEPAQ 240 KLKTAKRRRA AEVHNLSERR RRDRINEKMK ALQELIPHCN KTDKASMLDE AIEYLRSLQL 300 QLQVMWMGGG MATAAPVMFP AGVHQYMQRM VGPPHVAAMP RLPPFMAPPA VQSPPVGQMP 360 VADPYAGCLA VDSLQTPGPM HYLQGVNFYQ QQRPAPPPPP VVPGGSLPAA TALTLPSDNT 420 LHKEHEAGDP HTCPVSVSTS GAVRVNCSEL NRRQCWQTCV PVSTLKKPGH GRSEQKGAAV 480 TLSSHPLSSR ELLPRAPATV ALVEQPLADR NNEACGENNF HQA |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 178 | 186 | ARARDKRKQ |
| 2 | 181 | 187 | RDKRKQR |
| 3 | 241 | 247 | LKTAKRR |
| 4 | 246 | 261 | RRRAAEVHNLSERRRR |
| 5 | 257 | 262 | ERRRRD |
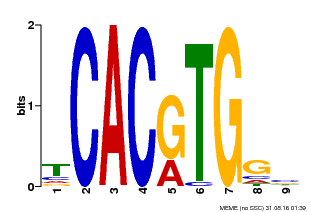
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00606 | ChIP-seq | Transfer from AT2G43010 | Download |
| |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_004963135.1 | 1e-141 | transcription factor PIF4 isoform X1 | ||||
| Refseq | XP_004963136.1 | 1e-141 | transcription factor PIF4 isoform X1 | ||||
| Refseq | XP_022680515.1 | 1e-141 | transcription factor PIF4 isoform X1 | ||||
| TrEMBL | K3Z6M7 | 1e-140 | K3Z6M7_SETIT; Uncharacterized protein | ||||
| STRING | Si022196m | 1e-140 | (Setaria italica) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP3107 | 36 | 77 |



