 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Zmw_sc03476.1.g00010.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; PACMAD clade; Chloridoideae; Zoysieae; Zoysiinae; Zoysia
|
||||||||
| Family | CPP | ||||||||
| Protein Properties | Length: 356aa MW: 39548.6 Da PI: 8.2852 | ||||||||
| Description | CPP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | TCR | 45 | 2.1e-14 | 67 | 104 | 4 | 41 |
TCR 4 kgCnCkkskClkkYCeCfaagkkCseeCkCedCkNkee 41
k C CkkskClk+YC Cf +g++Cse+C C+ C+Nk++
Zmw_sc03476.1.g00010.1.sm.mkhc 67 KYCACKKSKCLKLYCPCFTDGSYCSEKCGCQPCFNKDS 104
78*********************************975 PP
| |||||||
| 2 | TCR | 48.3 | 2e-15 | 151 | 187 | 3 | 39 |
TCR 3 kkgCnCkkskClkkYCeCfaagkkCseeCkCedCkNk 39
++gCnCkks+ClkkYC+C+++g+ Cs C+Ce C+N
Zmw_sc03476.1.g00010.1.sm.mkhc 151 RRGCNCKKSSCLKKYCDCYQEGTGCSLFCRCEACQNP 187
589*********************************6 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SMART | SM01114 | 1.4E-11 | 64 | 105 | IPR033467 | Tesmin/TSO1-like CXC domain |
| PROSITE profile | PS51634 | 29.266 | 65 | 189 | IPR005172 | CRC domain |
| Pfam | PF03638 | 1.1E-10 | 67 | 102 | IPR005172 | CRC domain |
| SMART | SM01114 | 5.3E-14 | 149 | 190 | IPR033467 | Tesmin/TSO1-like CXC domain |
| Pfam | PF03638 | 8.5E-11 | 152 | 186 | IPR005172 | CRC domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009934 | Biological Process | regulation of meristem structural organization | ||||
| GO:0048444 | Biological Process | floral organ morphogenesis | ||||
| GO:0051302 | Biological Process | regulation of cell division | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 356 aa Download sequence Send to blast |
MEATPISMKP PSPAAVATPP TAVEPQFSPQ PAAGAAEAEP GGMNQLVLTS NVKRQRVDEA 60 ADGNGFKYCA CKKSKCLKLY CPCFTDGSYC SEKCGCQPCF NKDSYAETVQ ATRKVLLSRQ 120 KRMSLKINRR SETNAEAMED AHNSSSSTPP RRGCNCKKSS CLKKYCDCYQ EGTGCSLFCR 180 CEACQNPFGK NEGIMAEDSR RYLYTGADLD HSEGEHEFVV ERSPRLQSPI SKESSFQTPP 240 HLRASSRDTH AFPQALSQWQ PLPRSGHCSK RNSNDRVMDD SGNYKYSSHG WLLSKHEDSY 300 SISKCIQILN GMVELSQVEK SVAPDVFLQQ GNREIFISLG GEVRALWLKR KIQHLT |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 5fd3_A | 3e-11 | 67 | 186 | 12 | 120 | Protein lin-54 homolog |
| 5fd3_B | 3e-11 | 67 | 186 | 12 | 120 | Protein lin-54 homolog |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00370 | DAP | Transfer from AT3G22780 | Download |
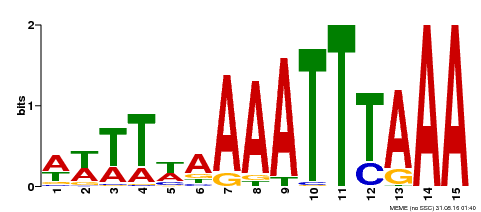
| |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_025810095.1 | 0.0 | protein tesmin/TSO1-like CXC 3 | ||||
| TrEMBL | A0A0A8ZWM0 | 0.0 | A0A0A8ZWM0_ARUDO; Uncharacterized protein | ||||
| STRING | Pavir.Ca01393.1.p | 0.0 | (Panicum virgatum) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP10159 | 29 | 37 |



