 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Zmw_sc03662.1.g00060.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; PACMAD clade; Chloridoideae; Zoysieae; Zoysiinae; Zoysia
|
||||||||
| Family | bHLH | ||||||||
| Protein Properties | Length: 260aa MW: 28628.5 Da PI: 9.6947 | ||||||||
| Description | bHLH family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | HLH | 41 | 3.5e-13 | 83 | 128 | 4 | 54 |
HHHHHHHHHHHHHHHHHHHHHCTSCCC...TTS-STCHHHHHHHHHHHHHH CS
HLH 4 ahnerErrRRdriNsafeeLrellPkaskapskKlsKaeiLekAveYIksL 54
+h e+Er+RR+++N++f Lr ++P+ K++Ka+ L A+ YI++L
Zmw_sc03662.1.g00060.1.es.mkhc 83 NHVEAERQRREKLNQRFYALRAVVPNV-----SKMDKASLLGDAISYINEL 128
799***********************6.....5****************98 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS50888 | 17.39 | 79 | 128 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SuperFamily | SSF47459 | 9.55E-19 | 82 | 153 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| CDD | cd00083 | 1.17E-12 | 82 | 132 | No hit | No description |
| Pfam | PF00010 | 1.2E-10 | 83 | 128 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene3D | G3DSA:4.10.280.10 | 6.3E-19 | 83 | 150 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SMART | SM00353 | 5.6E-16 | 85 | 134 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006952 | Biological Process | defense response | ||||
| GO:0009718 | Biological Process | anthocyanin-containing compound biosynthetic process | ||||
| GO:0009753 | Biological Process | response to jasmonic acid | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0043425 | Molecular Function | bHLH transcription factor binding | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 260 aa Download sequence Send to blast |
MEATSRASNT NLHPAATANE GMLSFSSAPT TRPAKSESDH SDLDASVREV ESSRVVAPPP 60 PEAEKRPRKR GRKPANGREE PLNHVEAERQ RREKLNQRFY ALRAVVPNVS KMDKASLLGD 120 AISYINELRG KLTALESDKE TMQSQIEALK KERDARPAPQ AGGFGHDAGP RCHAVEIDAK 180 ILGLEAMIRV QCHKRNHPSA RLMTALRELD LDVCHASVSV VKDLMIQQVA VKMASRIYSQ 240 DQLNAVLYSR LAEPGSVVGR |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 5gnj_A | 4e-35 | 77 | 156 | 2 | 81 | Transcription factor MYC2 |
| 5gnj_B | 4e-35 | 77 | 156 | 2 | 81 | Transcription factor MYC2 |
| 5gnj_E | 4e-35 | 77 | 156 | 2 | 81 | Transcription factor MYC2 |
| 5gnj_F | 4e-35 | 77 | 156 | 2 | 81 | Transcription factor MYC2 |
| 5gnj_G | 4e-35 | 77 | 156 | 2 | 81 | Transcription factor MYC2 |
| 5gnj_I | 4e-35 | 77 | 156 | 2 | 81 | Transcription factor MYC2 |
| 5gnj_M | 4e-35 | 77 | 156 | 2 | 81 | Transcription factor MYC2 |
| 5gnj_N | 4e-35 | 77 | 156 | 2 | 81 | Transcription factor MYC2 |
| Search in ModeBase | ||||||
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 64 | 72 | KRPRKRGRK |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcriptional activator involved in jasmonate (JA) signaling pathway during spikelet development. Binds to the G2 region G-box (5'-CACGTG-3') of the MADS1 promoter and thus directly regulates the expression of MADS1. Its function in MADS1 activation is abolished by TIFY3/JAZ1 which directly target MYC2 during spikelet development. {ECO:0000269|PubMed:24647160}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00085 | PBM | Transfer from AT5G46760 | Download |
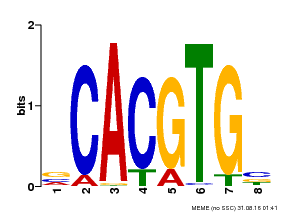
| |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_012698381.1 | 1e-167 | LOW QUALITY PROTEIN: transcription factor MYC2-like | ||||
| Swissprot | Q336P5 | 1e-155 | MYC2_ORYSJ; Transcription factor MYC2 | ||||
| TrEMBL | A0A1E5W733 | 1e-166 | A0A1E5W733_9POAL; Transcription factor MYC2 | ||||
| STRING | Pavir.J38781.1.p | 1e-170 | (Panicum virgatum) | ||||
| STRING | Si039973m | 1e-167 | (Setaria italica) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP4452 | 31 | 57 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



