 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Zmw_sc05892.1.g00030.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; PACMAD clade; Chloridoideae; Zoysieae; Zoysiinae; Zoysia
|
||||||||
| Family | G2-like | ||||||||
| Protein Properties | Length: 138aa MW: 14982.7 Da PI: 9.5444 | ||||||||
| Description | G2-like family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | G2-like | 98.9 | 3.6e-31 | 71 | 124 | 2 | 55 |
G2-like 2 prlrWtpeLHerFveaveqLGGsekAtPktilelmkvkgLtlehvkSHLQkYRl 55
prlrWtpeLH rF++ave+LGG+++AtPk +l lm+vkgL++ hvkSHLQ+YR+
Zmw_sc05892.1.g00030.1.sm.mk 71 PRLRWTPELHLRFLRAVERLGGQDRATPKLVLLLMNVKGLSIGHVKSHLQMYRS 124
9****************************************************7 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:1.10.10.60 | 1.9E-29 | 67 | 125 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS51294 | 12.951 | 67 | 127 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 1.08E-15 | 68 | 125 | IPR009057 | Homeodomain-like |
| TIGRFAMs | TIGR01557 | 1.7E-23 | 70 | 126 | IPR006447 | Myb domain, plants |
| Pfam | PF00249 | 9.3E-10 | 72 | 123 | IPR001005 | SANT/Myb domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 138 aa Download sequence Send to blast |
MEEISSKSFA SSKNQGDPEE GDAAADAPHY CHEGSSSHSS SVDMEVAAGG DSSRKTAASS 60 SVRPYVRSKN PRLRWTPELH LRFLRAVERL GGQDRATPKL VLLLMNVKGL SIGHVKSHLQ 120 MYRSKKIDDS GQGKSTDS |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 6j4r_A | 2e-18 | 72 | 126 | 3 | 57 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_B | 2e-18 | 72 | 126 | 3 | 57 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_C | 2e-18 | 72 | 126 | 3 | 57 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_D | 2e-18 | 72 | 126 | 3 | 57 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00297 | DAP | Transfer from AT2G38300 | Download |
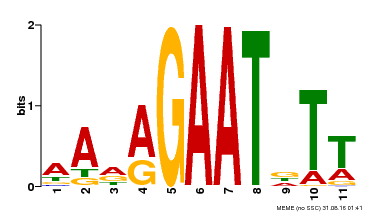
| |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_021317375.1 | 6e-53 | putative Myb family transcription factor At1g14600 | ||||
| Refseq | XP_021317376.1 | 6e-53 | putative Myb family transcription factor At1g14600 | ||||
| Refseq | XP_021321607.1 | 3e-50 | uncharacterized protein LOC8064764 | ||||
| TrEMBL | C5YQ15 | 8e-49 | C5YQ15_SORBI; Uncharacterized protein | ||||
| STRING | Sb08g000510.1 | 1e-50 | (Sorghum bicolor) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP4408 | 36 | 71 |



