 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Zosma127g00350.1 | ||||||||
| Common Name | ZOSMA_127G00350 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Alismatales; Zosteraceae; Zostera
|
||||||||
| Family | MYB | ||||||||
| Protein Properties | Length: 252aa MW: 28964.4 Da PI: 7.6135 | ||||||||
| Description | MYB family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 53 | 8e-17 | 13 | 60 | 1 | 48 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqkyl 48
+g+WT+eEd +lv v ++G ++W+ Ia+ g++Rt+k+c++rw +yl
Zosma127g00350.1 13 KGPWTEEEDMQLVYFVSLFGERRWDFIAKVSGLHRTGKSCRLRWVNYL 60
79********************************************97 PP
| |||||||
| 2 | Myb_DNA-binding | 53.5 | 5.3e-17 | 66 | 111 | 1 | 48 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqkyl 48
rgr++++E+ l ++++ ++G++ W++Iar+++ gRt++++k++w++++
Zosma127g00350.1 66 RGRMSPQEERLVLELHSRWGNR-WSRIARKLP-GRTDNEIKNYWRTHM 111
89********************.*********.************986 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51294 | 16.598 | 8 | 60 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 4.03E-30 | 11 | 107 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 3.5E-16 | 12 | 62 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 9.5E-15 | 13 | 60 | IPR001005 | SANT/Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 5.0E-20 | 14 | 67 | IPR009057 | Homeodomain-like |
| CDD | cd00167 | 3.57E-11 | 15 | 60 | No hit | No description |
| PROSITE profile | PS51294 | 25.948 | 61 | 115 | IPR017930 | Myb domain |
| SMART | SM00717 | 4.4E-16 | 65 | 113 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 5.0E-15 | 66 | 110 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 2.27E-10 | 68 | 111 | No hit | No description |
| Gene3D | G3DSA:1.10.10.60 | 3.5E-23 | 68 | 114 | IPR009057 | Homeodomain-like |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009723 | Biological Process | response to ethylene | ||||
| GO:0009739 | Biological Process | response to gibberellin | ||||
| GO:0009751 | Biological Process | response to salicylic acid | ||||
| GO:0009753 | Biological Process | response to jasmonic acid | ||||
| GO:0010200 | Biological Process | response to chitin | ||||
| GO:0046686 | Biological Process | response to cadmium ion | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 252 aa Download sequence Send to blast |
MEEKNLSSTT MRKGPWTEEE DMQLVYFVSL FGERRWDFIA KVSGLHRTGK SCRLRWVNYL 60 HPGLKRGRMS PQEERLVLEL HSRWGNRWSR IARKLPGRTD NEIKNYWRTH MRKKAQERKR 120 NIAASTASSS SSTITTTHAV GESGGGGGNQ TLVLQPRETT ETGCYPMEKI WDEIDQSSEM 180 MSRPTSVVDY HFKEENWRID EHSCFHPPTT SSYDDVEGGL WRMNQEEEKE LMNLMSSIGC 240 GGDTLLSNPN T* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1gv2_A | 5e-29 | 13 | 115 | 4 | 105 | MYB PROTO-ONCOGENE PROTEIN |
| 1h88_C | 2e-28 | 13 | 115 | 58 | 159 | MYB PROTO-ONCOGENE PROTEIN |
| 1h89_C | 2e-28 | 13 | 115 | 58 | 159 | MYB PROTO-ONCOGENE PROTEIN |
| 1mse_C | 6e-29 | 13 | 115 | 4 | 105 | C-Myb DNA-Binding Domain |
| 1msf_C | 6e-29 | 13 | 115 | 4 | 105 | C-Myb DNA-Binding Domain |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor. {ECO:0000305}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00600 | PBM | Transfer from AT5G59780 | Download |
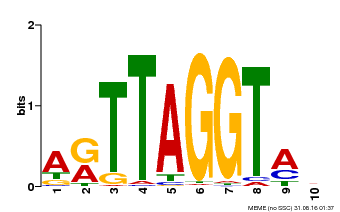
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Isoform MYB59-1 is induced by jasmonate (JA), salicylic acid (SA), gibberellic acid (GA), and ethylene. Also induced by cadmium (Cd). {ECO:0000269|PubMed:16463103, ECO:0000269|PubMed:16531467}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_006400918.1 | 8e-81 | transcription factor MYB59 isoform X1 | ||||
| Swissprot | Q4JL84 | 5e-78 | MYB59_ARATH; Transcription factor MYB59 | ||||
| TrEMBL | A0A0K9Q006 | 0.0 | A0A0K9Q006_ZOSMR; Transcription factor MYB59 | ||||
| STRING | XP_006400918.1 | 3e-80 | (Eutrema salsugineum) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP636 | 37 | 169 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G59780.3 | 1e-77 | myb domain protein 59 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Zosma127g00350.1 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



