 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Zosma15g00510.1 | ||||||||
| Common Name | ZOSMA_15G00510 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Alismatales; Zosteraceae; Zostera
|
||||||||
| Family | LBD | ||||||||
| Protein Properties | Length: 210aa MW: 22747.6 Da PI: 7.4316 | ||||||||
| Description | LBD family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | DUF260 | 137.9 | 3.6e-43 | 30 | 128 | 1 | 99 |
DUF260 1 aCaaCkvlrrkCakdCvlapyfpaeqpkkfanvhklFGasnvlkllkalpeeeredamsslvyeAearardPvyGavgvilklqqqleqlkael 94
+CaaCk+lrrkC ++C++apyfp ++p+kfanvhk+FGasn++kll++l + +reda++sl+yeAear++dPvyG+vg i+ lq+q+++l++el
Zosma15g00510.1 30 PCAACKLLRRKCIPGCIFAPYFPPDDPQKFANVHKIFGASNITKLLNDLLPYQREDAVNSLAYEAEARMKDPVYGCVGAISVLQKQVQRLQNEL 123
7********************************************************************************************* PP
DUF260 95 allke 99
+++++
Zosma15g00510.1 124 DAANT 128
99876 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS50891 | 26.955 | 29 | 130 | IPR004883 | Lateral organ boundaries, LOB |
| Pfam | PF03195 | 1.6E-42 | 30 | 127 | IPR004883 | Lateral organ boundaries, LOB |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0010199 | Biological Process | organ boundary specification between lateral organs and the meristem | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 210 aa Download sequence Send to blast |
MAQAKNNNSC TRKNIMASSQ SNSSTNSTPP CAACKLLRRK CIPGCIFAPY FPPDDPQKFA 60 NVHKIFGASN ITKLLNDLLP YQREDAVNSL AYEAEARMKD PVYGCVGAIS VLQKQVQRLQ 120 NELDAANTNL LRYACNIVDI PSSMDRLPLM GSTSSTGLGE GSVGGGDLYH HHHASGLPIP 180 YTQAWGNNNR SNYHPPGDMD EMDGGERSM* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 5ly0_A | 5e-66 | 30 | 144 | 11 | 126 | LOB family transfactor Ramosa2.1 |
| 5ly0_B | 5e-66 | 30 | 144 | 11 | 126 | LOB family transfactor Ramosa2.1 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Not known; ectopic expression of LOB leads to alterations in the size and shape of leaves and floral organs and causes male and female sterility. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00581 | DAP | Transfer from AT5G63090 | Download |
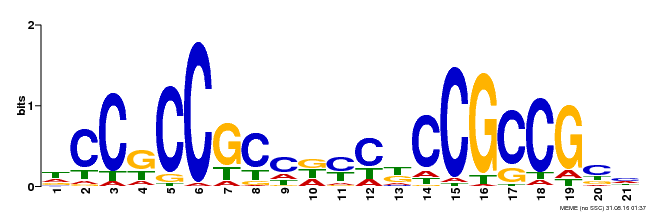
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Positively regulated within the shoot apex by both ASYMMETRIC LEAVES 1 (AS1) and ASYMMETRIC LEAVES 2 (AS2/LBD6) and by KNAT1. {ECO:0000269|PubMed:11934861}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_017699393.2 | 2e-79 | LOW QUALITY PROTEIN: LOB domain-containing protein 25-like | ||||
| Swissprot | Q9FML4 | 5e-62 | LOB_ARATH; Protein LATERAL ORGAN BOUNDARIES | ||||
| TrEMBL | A0A0K9PUJ9 | 1e-156 | A0A0K9PUJ9_ZOSMR; LOB domain protein 1 | ||||
| STRING | XP_008795982.1 | 5e-79 | (Phoenix dactylifera) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP8813 | 35 | 45 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G63090.4 | 1e-62 | LBD family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Zosma15g00510.1 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



