 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Zosma19g00170.1 | ||||||||
| Common Name | ZOSMA_19G00170 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Alismatales; Zosteraceae; Zostera
|
||||||||
| Family | C2H2 | ||||||||
| Protein Properties | Length: 596aa MW: 64338.2 Da PI: 9.6251 | ||||||||
| Description | C2H2 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | zf-C2H2 | 15.3 | 5.9e-05 | 84 | 106 | 1 | 23 |
EEETTTTEEESSHHHHHHHHHHT CS
zf-C2H2 1 ykCpdCgksFsrksnLkrHirtH 23
+ C+ C+k F r nL+ H r H
Zosma19g00170.1 84 FICEVCKKGFQREQNLQLHRRGH 106
89*******************88 PP
| |||||||
| 2 | zf-C2H2 | 12.5 | 0.00045 | 160 | 182 | 1 | 23 |
EEETTTTEEESSHHHHHHHHHHT CS
zf-C2H2 1 ykCpdCgksFsrksnLkrHirtH 23
+kC +C+k++ +s+ k H +t+
Zosma19g00170.1 160 WKCDKCSKRYAVQSDWKAHSKTC 182
58*****************9998 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:3.30.160.60 | 3.9E-5 | 83 | 106 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| SuperFamily | SSF57667 | 3.2E-7 | 83 | 106 | No hit | No description |
| PROSITE profile | PS50157 | 10.679 | 84 | 106 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 0.012 | 84 | 106 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE pattern | PS00028 | 0 | 86 | 106 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 190 | 125 | 155 | IPR015880 | Zinc finger, C2H2-like |
| Gene3D | G3DSA:3.30.160.60 | 1.1E-4 | 148 | 181 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| SuperFamily | SSF57667 | 3.2E-7 | 155 | 180 | No hit | No description |
| SMART | SM00355 | 140 | 160 | 180 | IPR015880 | Zinc finger, C2H2-like |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0003676 | Molecular Function | nucleic acid binding | ||||
| GO:0046872 | Molecular Function | metal ion binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 596 aa Download sequence Send to blast |
MAAPSSSSPF FGIREDDLRT NIHTHMKQQQ ISEMVGPGPA SSTGGAPPSG PLMAKKKRNL 60 PGTPNPDAEV VALSPKTLMA TNRFICEVCK KGFQREQNLQ LHRRGHNLPW KLRQKSTKEV 120 KRKVYLCPEP TCVHHDRSRA LGDLTGIKKH FSRKHGEKKW KCDKCSKRYA VQSDWKAHSK 180 TCGTREYRCD CGTLFSRRDS FITHRAFCDA LAQESARIPT GMGINSTYLY LNNGNGNGNG 240 NGISTVSLAN INQMNTQRDQ NHASNDVLRL GSRSGSQFGH LVGSNSPTVQ SGPSSNGFFF 300 GQHHQQDHHF QDDHPLLHNK PFHGGLMQPP DLQASSSNAH AGATNLFNLG FFSNSTRSSS 360 SSQSNHLLMS DQFNSGSGSV NNEQGTLFSG SLIGAGGGDN NIYSTSLQND SLLPQMSATA 420 LLQKAAQMGS TTSAGITPSS LPRSTKPSEL SMTHFAGTGD HGSIRSQIEN ENNFQDLMNS 480 LANGNAASMF GVANSGTTTA VFGNGGYEPN RTLGFDGYSS NFGEVTDDTK LHQSMNPARN 540 MAGGGSDGLT RDFLGVAGMM RRDQQHNHHG LDIGSFDSKL KSNTNTRTFQ GGNLQ* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 5b3h_C | 2e-34 | 156 | 220 | 3 | 67 | Zinc finger protein JACKDAW |
| 5b3h_F | 2e-34 | 156 | 220 | 3 | 67 | Zinc finger protein JACKDAW |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor acting as a positive regulator of the starch synthase SS4. Controls chloroplast development and starch granule formation (PubMed:22898356). Binds DNA via its zinc fingers (PubMed:24821766). Recognizes and binds to SCL3 promoter sequence 5'-AGACAA-3' to promotes its expression when in complex with RGA (PubMed:24821766). {ECO:0000269|PubMed:22898356, ECO:0000269|PubMed:24821766}. | |||||
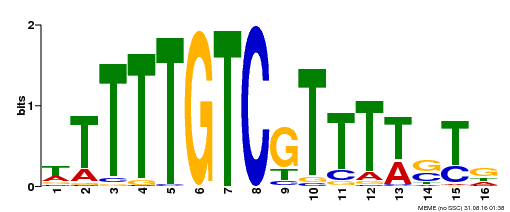
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00255 | DAP | Transfer from AT2G02070 | Download |
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010270755.1 | 0.0 | PREDICTED: protein indeterminate-domain 5, chloroplastic-like | ||||
| Refseq | XP_010270757.1 | 0.0 | PREDICTED: protein indeterminate-domain 5, chloroplastic-like | ||||
| Swissprot | Q9ZUL3 | 1e-120 | IDD5_ARATH; Protein indeterminate-domain 5, chloroplastic | ||||
| TrEMBL | A0A0K9PQB0 | 0.0 | A0A0K9PQB0_ZOSMR; Putative Zinc finger protein | ||||
| STRING | XP_010270754.1 | 0.0 | (Nelumbo nucifera) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP1733 | 37 | 98 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G02080.1 | 1e-113 | indeterminate(ID)-domain 4 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Zosma19g00170.1 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



