 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Zosma239g00280.1 | ||||||||
| Common Name | ZOSMA_239G00280 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Alismatales; Zosteraceae; Zostera
|
||||||||
| Family | MYB | ||||||||
| Protein Properties | Length: 375aa MW: 40738.3 Da PI: 6.19 | ||||||||
| Description | MYB family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 52.3 | 1.3e-16 | 55 | 100 | 1 | 47 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHH CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqky 47
+g+W++eEd +l ++v ++G ++W++Iar + gR++k+c++rw +
Zosma239g00280.1 55 KGPWSPEEDAILSRLVGKFGARNWSLIARGIS-GRSGKSCRLRWCNQ 100
79******************************.***********985 PP
| |||||||
| 2 | Myb_DNA-binding | 55.2 | 1.6e-17 | 107 | 151 | 1 | 47 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHH CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqky 47
r++++ eEd++++ a++ +G++ W++Iar + gRt++ +k++w++
Zosma239g00280.1 107 RKPFSDEEDQIIIAAHAIHGNK-WASIARLLE-GRTDNAIKNHWNST 151
789*******************.*********.***********986 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51294 | 20.05 | 50 | 101 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 1.01E-29 | 52 | 148 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 2.4E-14 | 54 | 103 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 1.2E-15 | 55 | 100 | IPR001005 | SANT/Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 4.5E-25 | 56 | 108 | IPR009057 | Homeodomain-like |
| CDD | cd00167 | 8.47E-14 | 57 | 99 | No hit | No description |
| PROSITE profile | PS51294 | 26.28 | 102 | 156 | IPR017930 | Myb domain |
| SMART | SM00717 | 1.5E-16 | 106 | 154 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 8.3E-15 | 107 | 151 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 2.18E-12 | 109 | 152 | No hit | No description |
| Gene3D | G3DSA:1.10.10.60 | 5.4E-23 | 109 | 155 | IPR009057 | Homeodomain-like |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009751 | Biological Process | response to salicylic acid | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 375 aa Download sequence Send to blast |
MENEQFSVPA ADAAVESGGG GYSGGVHEVE PEEDGTGEEV MGGAGRGGGG MDRVKGPWSP 60 EEDAILSRLV GKFGARNWSL IARGISGRSG KSCRLRWCNQ LDPCVKRKPF SDEEDQIIIA 120 AHAIHGNKWA SIARLLEGRT DNAIKNHWNS TLKRQCFEFD RPTIHVMKGD VPPESGSADR 180 KGSSEETLSC ADANSARFPD RRDGSSPETM SNHQFEDRTH MRENKDPPNL FRPIARLSAF 240 RPVSFSNANV SVVPNSAPLC CPSIQFMMKH DNSEQIHGEP PQVPARCGYG CCRSSTHQNP 300 SSLLGPEFVE FEDLPSISSH DIASIATELS SVAWLKSGLD SSSTKLLDWS VPNGMTSQGQ 360 HTAPHFPQLN NRMV* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1a5j_A | 9e-39 | 52 | 155 | 4 | 107 | B-MYB |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00337 | DAP | Transfer from AT3G09230 | Download |
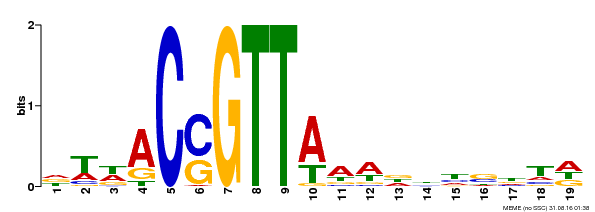
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010262707.1 | 1e-118 | PREDICTED: uncharacterized protein LOC104601164 | ||||
| TrEMBL | A0A0K9PHR5 | 0.0 | A0A0K9PHR5_ZOSMR; Uncharacterized protein | ||||
| STRING | XP_010262707.1 | 1e-118 | (Nelumbo nucifera) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP8990 | 36 | 47 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G09230.1 | 4e-80 | myb domain protein 1 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Zosma239g00280.1 |



