 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Zosma248g00110.1 | ||||||||
| Common Name | ZOSMA_248G00110 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Alismatales; Zosteraceae; Zostera
|
||||||||
| Family | HD-ZIP | ||||||||
| Protein Properties | Length: 304aa MW: 34523.8 Da PI: 6.4039 | ||||||||
| Description | HD-ZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Homeobox | 54.9 | 1.5e-17 | 92 | 145 | 3 | 56 |
--SS--HHHHHHHHHHHHHSSS--HHHHHHHHHHCTS-HHHHHHHHHHHHHHHH CS
Homeobox 3 kRttftkeqleeLeelFeknrypsaeereeLAkklgLterqVkvWFqNrRakek 56
k+++++ eq+++Le+ Fe +++ e++ +LA+ lgL+ rq+ +WFqNrRa++k
Zosma248g00110.1 92 KKRRLNMEQVRTLEKNFELGNKLEPERKLQLARALGLQPRQIAIWFQNRRARWK 145
456899***********************************************9 PP
| |||||||
| 2 | HD-ZIP_I/II | 129.4 | 1.4e-41 | 91 | 183 | 1 | 93 |
HD-ZIP_I/II 1 ekkrrlskeqvklLEesFeeeekLeperKvelareLglqprqvavWFqnrRARtktkqlEkdyeaLkraydalkeenerLekeveeLreelke 93
ekkrrl+ eqv++LE++Fe +kLeperK +lar+Lglqprq+a+WFqnrRAR+ktkqlEkdy++Lk+++da+k+en+ L++++++Lr+e+++
Zosma248g00110.1 91 EKKRRLNMEQVRTLEKNFELGNKLEPERKLQLARALGLQPRQIAIWFQNRRARWKTKQLEKDYDVLKKKFDAVKSENDLLQTQNKKLRAEIMA 183
69**************************************************************************************99975 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF46689 | 2.31E-19 | 73 | 149 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS50071 | 17.006 | 87 | 147 | IPR001356 | Homeobox domain |
| SMART | SM00389 | 1.6E-17 | 90 | 151 | IPR001356 | Homeobox domain |
| Pfam | PF00046 | 5.4E-15 | 92 | 145 | IPR001356 | Homeobox domain |
| CDD | cd00086 | 1.68E-15 | 92 | 148 | No hit | No description |
| Gene3D | G3DSA:1.10.10.60 | 1.5E-18 | 94 | 155 | IPR009057 | Homeodomain-like |
| PRINTS | PR00031 | 1.7E-5 | 118 | 127 | IPR000047 | Helix-turn-helix motif |
| PROSITE pattern | PS00027 | 0 | 122 | 145 | IPR017970 | Homeobox, conserved site |
| PRINTS | PR00031 | 1.7E-5 | 127 | 143 | IPR000047 | Helix-turn-helix motif |
| Pfam | PF02183 | 2.4E-14 | 147 | 187 | IPR003106 | Leucine zipper, homeobox-associated |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009744 | Biological Process | response to sucrose | ||||
| GO:0048826 | Biological Process | cotyledon morphogenesis | ||||
| GO:0080022 | Biological Process | primary root development | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 304 aa Download sequence Send to blast |
MGCNGMASSF FPPTASFLLQ MQTPHDKSHE REAPRQNLPS SLAPIFSQDY GHGLLGKRSM 60 SLHSTMDQQQ VCHDEVNGED ELSDDCSQLG EKKRRLNMEQ VRTLEKNFEL GNKLEPERKL 120 QLARALGLQP RQIAIWFQNR RARWKTKQLE KDYDVLKKKF DAVKSENDLL QTQNKKLRAE 180 IMALNGREIS SEPINLNKET EVSCSYRSDN SSDINLEISR TPGKNESPMN NIAQQSMSFF 240 PITPVVPQPP SQVMVSSQLL QRQELQGPKV EHGGEDSFCN IFCSLDDQPA GFWAGWQEQQ 300 SFH* |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 139 | 147 | RRARWKTKQ |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor that may act in the sucrose-signaling pathway. {ECO:0000269|PubMed:11292072}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00225 | DAP | Transfer from AT1G69780 | Download |
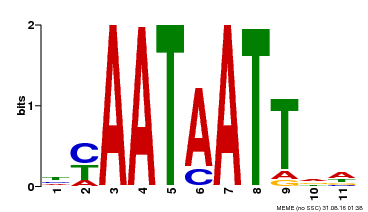
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_020275215.1 | 1e-112 | homeobox-leucine zipper protein HOX21-like | ||||
| Swissprot | Q8LC03 | 4e-86 | ATB13_ARATH; Homeobox-leucine zipper protein ATHB-13 | ||||
| TrEMBL | A0A0K9PIX8 | 0.0 | A0A0K9PIX8_ZOSMR; Homeobox-leucine zipper protein ATHB-13 | ||||
| STRING | XP_008797128.1 | 1e-106 | (Phoenix dactylifera) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP2289 | 38 | 94 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G69780.1 | 2e-85 | HD-ZIP family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Zosma248g00110.1 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



