 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Zosma31g01410.1 | ||||||||
| Common Name | ZOSMA_31G01410 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Alismatales; Zosteraceae; Zostera
|
||||||||
| Family | ERF | ||||||||
| Protein Properties | Length: 275aa MW: 30066.5 Da PI: 8.2738 | ||||||||
| Description | ERF family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | AP2 | 59.3 | 9.1e-19 | 100 | 150 | 2 | 56 |
AP2 2 gykGVrwdkkrgrWvAeIrdpsengkrkrfslgkfgtaeeAakaaiaarkklege 56
y+GVr + +g+Wv+eIr+p++ + r++lg+f tae+Aa+a++ a+++++ge
Zosma31g01410.1 100 TYRGVRMRN-WGKWVSEIREPKK---KSRIWLGTFPTAEMAARAHDVAALSIKGE 150
69****998.**********854...6************************9986 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:3.30.730.10 | 1.6E-31 | 100 | 158 | IPR001471 | AP2/ERF domain |
| PROSITE profile | PS51032 | 23.117 | 100 | 157 | IPR001471 | AP2/ERF domain |
| SuperFamily | SSF54171 | 2.16E-21 | 100 | 158 | IPR016177 | DNA-binding domain |
| SMART | SM00380 | 7.7E-37 | 100 | 163 | IPR001471 | AP2/ERF domain |
| Pfam | PF00847 | 2.2E-12 | 100 | 150 | IPR001471 | AP2/ERF domain |
| PRINTS | PR00367 | 1.0E-10 | 101 | 112 | IPR001471 | AP2/ERF domain |
| CDD | cd00018 | 1.95E-31 | 101 | 159 | No hit | No description |
| PRINTS | PR00367 | 1.0E-10 | 123 | 139 | IPR001471 | AP2/ERF domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0044212 | Molecular Function | transcription regulatory region DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 275 aa Download sequence Send to blast |
MEENRLKIEA CSSPRSPCSS NTTASSSITN CITGQKEYGF VISSGNSPSS SYSSCSLWLE 60 QRSGVKEMAT ERGRRDGLVG NESKMKNKRN RDGGGGDHPT YRGVRMRNWG KWVSEIREPK 120 KKSRIWLGTF PTAEMAARAH DVAALSIKGE STYLNFPELA SELPRPATSS PKDIQAAAAK 180 AASATFSQNR RVVGVVRTDK ENLASNSNSI NRSPILEIPA SSSCEEDKAL FDLPDLFVDH 240 CKNVSCFYSF LLSSTTDDAA GEISFRQESP FLWE* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 5wx9_A | 3e-18 | 93 | 158 | 7 | 73 | Ethylene-responsive transcription factor ERF096 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probably acts as a transcriptional activator. Binds to the GCC-box pathogenesis-related promoter element. May be involved in the regulation of gene expression by stress factors and by components of stress signal transduction pathways (By similarity). {ECO:0000250}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00312 | DAP | Transfer from AT2G44940 | Download |
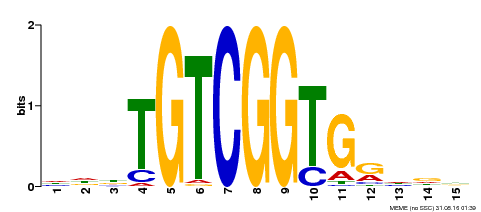
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_023757522.1 | 5e-60 | ethylene-responsive transcription factor ERF038 | ||||
| Swissprot | Q8LBQ7 | 8e-55 | ERF34_ARATH; Ethylene-responsive transcription factor ERF034 | ||||
| TrEMBL | A0A0K9P997 | 0.0 | A0A0K9P997_ZOSMR; Uncharacterized protein | ||||
| STRING | cassava4.1_021971m | 3e-59 | (Manihot esculenta) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP159 | 35 | 354 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G44940.1 | 7e-53 | ERF family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Zosma31g01410.1 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



