 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Zosma34g00290.1 | ||||||||
| Common Name | ZOSMA_34G00290 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Alismatales; Zosteraceae; Zostera
|
||||||||
| Family | WOX | ||||||||
| Protein Properties | Length: 323aa MW: 36354.5 Da PI: 5.7035 | ||||||||
| Description | WOX family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Homeobox | 59 | 7.6e-19 | 43 | 104 | 2 | 57 |
T--SS--HHHHHHHHHHHHH..SSS--HHHHHHHHHHC....TS-HHHHHHHHHHHHHHHHC CS
Homeobox 2 rkRttftkeqleeLeelFek..nrypsaeereeLAkkl....gLterqVkvWFqNrRakekk 57
++R+ +t++q+++L++l+++ r p+a+++++++++l +++ ++V++WFqN++a+e++
Zosma34g00290.1 43 STRWIPTTDQIRILKDLYYNsgLRSPNADQIQRISARLrqygKIEGKNVFYWFQNHKARERQ 104
58*****************8789*************************************97 PP
| |||||||
| 2 | Wus_type_Homeobox | 112.5 | 2.3e-36 | 42 | 106 | 2 | 65 |
Wus_type_Homeobox 2 artRWtPtpeQikiLeelyk.sGlrtPnkeeiqritaeLeeyGkiedkNVfyWFQNrkaRerqkq 65
++tRW Pt++Qi+iL++ly+ sGlr+Pn+++iqri+a+L++yGkie+kNVfyWFQN+kaRerqk+
Zosma34g00290.1 42 SSTRWIPTTDQIRILKDLYYnSGLRSPNADQIQRISARLRQYGKIEGKNVFYWFQNHKARERQKK 106
789***************9779*****************************************96 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF46689 | 5.56E-11 | 36 | 109 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS50071 | 9.863 | 39 | 105 | IPR001356 | Homeobox domain |
| SMART | SM00389 | 2.0E-4 | 41 | 109 | IPR001356 | Homeobox domain |
| CDD | cd00086 | 1.01E-4 | 44 | 106 | No hit | No description |
| Pfam | PF00046 | 3.5E-16 | 44 | 104 | IPR001356 | Homeobox domain |
| Gene3D | G3DSA:1.10.10.60 | 6.6E-6 | 45 | 104 | IPR009057 | Homeodomain-like |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0010492 | Biological Process | maintenance of shoot apical meristem identity | ||||
| GO:0080166 | Biological Process | stomium development | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 323 aa Download sequence Send to blast |
MEHHHQQQFH GGQPQLMSEN EEMNNNGSGG GSSKSSFLCR QSSTRWIPTT DQIRILKDLY 60 YNSGLRSPNA DQIQRISARL RQYGKIEGKN VFYWFQNHKA RERQKKRLTV DIAAAATTNK 120 FSNNTNMSSS SSPSGISPAS HAVYAGLGQM GSNGGGSVVM ERTSYMECSL PGGMPMPVPE 180 PNVGVGVGWV SVETGATTMQ QFYYHQQRPT TPFPAPHHHH HYYDDGYYSN NNKLQAHHLH 240 IREENQHPED DDEEEEEEEE DEEIETLPLF PIHSQETQSY FLDQQHAIHN QESNNMETSL 300 ELTLNSYYAA SSSSTPHGSE HD* |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00265 | DAP | Transfer from AT2G17950 | Download |
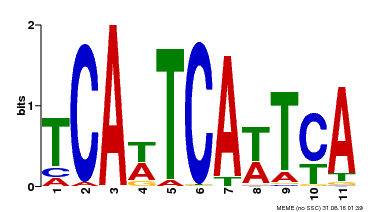
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| TrEMBL | A0A0K9P935 | 0.0 | A0A0K9P935_ZOSMR; WUSCHEL-related homeobox | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP12465 | 31 | 36 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G17950.1 | 2e-38 | WOX family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Zosma34g00290.1 |



