 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Zosma37g01300.1 | ||||||||
| Common Name | ZOSMA_37G01300 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Alismatales; Zosteraceae; Zostera
|
||||||||
| Family | C2H2 | ||||||||
| Protein Properties | Length: 251aa MW: 27206.9 Da PI: 8.873 | ||||||||
| Description | C2H2 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | zf-C2H2 | 13.6 | 0.00019 | 100 | 122 | 1 | 23 |
EEETTTTEEESSHHHHHHHHHHT CS
zf-C2H2 1 ykCpdCgksFsrksnLkrHirtH 23
y+C+ C+ksF++ L H +H
Zosma37g01300.1 100 YQCTTCDKSFPSFQALGGHRTSH 122
9***********99998888777 PP
| |||||||
| 2 | zf-C2H2 | 14.4 | 0.00011 | 169 | 191 | 1 | 23 |
EEETTTTEEESSHHHHHHHHHHT CS
zf-C2H2 1 ykCpdCgksFsrksnLkrHirtH 23
++C++Cg Fs+ L H+r+H
Zosma37g01300.1 169 HRCSICGSEFSSGQALGGHMRRH 191
79********************9 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF57667 | 4.59E-10 | 99 | 122 | No hit | No description |
| Pfam | PF13912 | 1.3E-12 | 99 | 124 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 0.019 | 100 | 122 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE profile | PS50157 | 10.907 | 100 | 127 | IPR007087 | Zinc finger, C2H2 |
| PROSITE pattern | PS00028 | 0 | 102 | 122 | IPR007087 | Zinc finger, C2H2 |
| SuperFamily | SSF57667 | 4.59E-10 | 164 | 191 | No hit | No description |
| Pfam | PF13912 | 1.5E-11 | 168 | 193 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 0.012 | 169 | 191 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE profile | PS50157 | 9.702 | 169 | 196 | IPR007087 | Zinc finger, C2H2 |
| PROSITE pattern | PS00028 | 0 | 171 | 191 | IPR007087 | Zinc finger, C2H2 |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0046872 | Molecular Function | metal ion binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 251 aa Download sequence Send to blast |
MGMEAVYDGS VDDYSSHSIM KGKRTKRRRN FNRSPMSTTT TTTTVSTTSS GSDISGSRNS 60 TDDTITKKEE EDEEMANCLI LLAQGRGLIT GDTDIGRCVY QCTTCDKSFP SFQALGGHRT 120 SHKKPKTSTS TAVMMIKGKN KSTQEESSLI KINNILLSPF VGGTKSKVHR CSICGSEFSS 180 GQALGGHMRR HRSTMVLEEK KEKKLVFGLD LNLPAPSEID MTVDPSSPSI GYHCILPILA 240 ELPLLSKLSS * |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00487 | DAP | Transfer from AT5G04390 | Download |
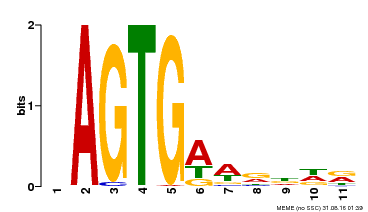
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_008779116.1 | 6e-48 | zinc finger protein ZAT5-like | ||||
| TrEMBL | A0A0K9P7U6 | 0.0 | A0A0K9P7U6_ZOSMR; Zinc finger family protein | ||||
| STRING | XP_008779116.1 | 2e-47 | (Phoenix dactylifera) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP2013 | 32 | 85 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G04390.1 | 4e-27 | C2H2 family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Zosma37g01300.1 |



