 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Zosma38g00570.1 | ||||||||
| Common Name | ZOSMA_38G00570 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Alismatales; Zosteraceae; Zostera
|
||||||||
| Family | MYB_related | ||||||||
| Protein Properties | Length: 632aa MW: 69022.7 Da PI: 5.8684 | ||||||||
| Description | MYB_related family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 36.9 | 8.5e-12 | 56 | 100 | 1 | 47 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHH CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqky 47
r +W+++E+e++++ +++ + Wk+I + +g +t q++s+ qky
Zosma38g00570.1 56 RESWSEQEHEKFIESLQLFDRD-WKKIEAFVG-SKTVIQIRSHAQKY 100
789*****************77.*********.*************9 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF46689 | 4.26E-15 | 50 | 106 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS51294 | 14.039 | 51 | 105 | IPR017930 | Myb domain |
| TIGRFAMs | TIGR01557 | 7.1E-16 | 54 | 103 | IPR006447 | Myb domain, plants |
| SMART | SM00717 | 5.2E-10 | 55 | 103 | IPR001005 | SANT/Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 1.1E-5 | 55 | 96 | IPR009057 | Homeodomain-like |
| Pfam | PF00249 | 1.8E-9 | 56 | 100 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 2.73E-7 | 58 | 101 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009651 | Biological Process | response to salt stress | ||||
| GO:0009723 | Biological Process | response to ethylene | ||||
| GO:0009733 | Biological Process | response to auxin | ||||
| GO:0009737 | Biological Process | response to abscisic acid | ||||
| GO:0009739 | Biological Process | response to gibberellin | ||||
| GO:0009751 | Biological Process | response to salicylic acid | ||||
| GO:0009753 | Biological Process | response to jasmonic acid | ||||
| GO:0032922 | Biological Process | circadian regulation of gene expression | ||||
| GO:0043966 | Biological Process | histone H3 acetylation | ||||
| GO:0046686 | Biological Process | response to cadmium ion | ||||
| GO:0048573 | Biological Process | photoperiodism, flowering | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 632 aa Download sequence Send to blast |
MLPVTTVGKD DGSGSFIAPA PVSGIGLSQN TEFVSDDATT TTTKKVRKPY TIKKTRESWS 60 EQEHEKFIES LQLFDRDWKK IEAFVGSKTV IQIRSHAQKY FLKIQKDGTN DHIPPPRPKR 120 KAAHPYPQKA SKNDTKSSQF SNPLQAPSYS VVSEQGYSFK RAVVDSSSIQ GTPNAITIAP 180 CWSQNTTTPI SIDNAPSWSQ NTTIPTISID DLPSWSQNTT IPTISIDDLP CWSHNTTTPI 240 SIDDLPCWSQ NTTKPISIDD FPGWSQNTTA PISIDDLPIS IDDFPGWSQN TTAPISIDDL 300 PISIDDLPIS IDDFPCWSQN TTTPISIDDL PCWSQNTTTP ISTNNISSGK DFTNDWRHVG 360 SSKNAPNKNF CSSSGKDNQG HVGSSKNAPN KNFCSSSGKD DWGHVGSSKN APNKNFYSSS 420 GKDDWGHVGS SKNAPNKNFC SSSGKDDWGH VGSSKNAPNK NFYSSSGKDD WGHVGSSKNA 480 PNKNFCSSSG KGLTDDWGYV GSSKNAPNKN CCSSSGKDDW GHVGSSKNAP NKNGCSSSGS 540 PSETWPISET SEQIRNIPAN RAIPDFVQVH QFIGSVLDPD TEDNHMETLK QMESIDVETV 600 LLLMKNLTVN LENPDFEEHR KLLSYYSDQM Q* |
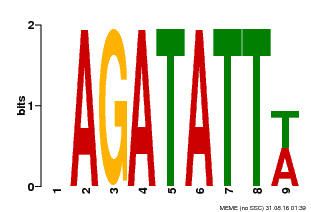
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00625 | PBM | Transfer from PK02532.1 | Download |
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | - | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| TrEMBL | A0A0K9P4L6 | 0.0 | A0A0K9P4L6_ZOSMR; Uncharacterized protein | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP1096 | 38 | 111 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G09600.2 | 9e-38 | MYB_related family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Zosma38g00570.1 |



