 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Zosma506g00020.1 | ||||||||
| Common Name | ZOSMA_506G00020 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Alismatales; Zosteraceae; Zostera
|
||||||||
| Family | MYB | ||||||||
| Protein Properties | Length: 484aa MW: 54070.9 Da PI: 8.4119 | ||||||||
| Description | MYB family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 57.8 | 2.5e-18 | 158 | 204 | 1 | 48 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqkyl 48
+g WT+eEd ll+ +vk++G + W+ I++++ gR +kqc++rw+++l
Zosma506g00020.1 158 KGQWTPEEDSLLISLVKEHGAKKWSDISKKLV-GRIGKQCRERWHNHL 204
799*****************************.*************97 PP
| |||||||
| 2 | Myb_DNA-binding | 55.2 | 1.6e-17 | 210 | 252 | 1 | 45 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHH CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwq 45
+++W+ eEd+llv+ +++lG++ W++Ia++++ gRt++ +k++w+
Zosma506g00020.1 210 KDNWSDEEDLLLVRSHRKLGNR-WAKIAKRIP-GRTENAIKNHWN 252
689*******************.*********.***********8 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51294 | 29.169 | 153 | 208 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 2.24E-30 | 156 | 251 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 7.5E-17 | 157 | 206 | IPR001005 | SANT/Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 6.7E-28 | 159 | 211 | IPR009057 | Homeodomain-like |
| Pfam | PF13921 | 2.4E-19 | 161 | 220 | No hit | No description |
| CDD | cd00167 | 6.91E-16 | 161 | 204 | No hit | No description |
| PROSITE profile | PS51294 | 21.801 | 209 | 259 | IPR017930 | Myb domain |
| SMART | SM00717 | 3.7E-17 | 209 | 257 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 4.77E-14 | 212 | 255 | No hit | No description |
| Gene3D | G3DSA:1.10.10.60 | 2.1E-20 | 212 | 257 | IPR009057 | Homeodomain-like |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0010439 | Biological Process | regulation of glucosinolate biosynthetic process | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:1904095 | Biological Process | negative regulation of endosperm development | ||||
| GO:2000692 | Biological Process | negative regulation of seed maturation | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 484 aa Download sequence Send to blast |
MQLILFLDSL ILHSSPTSFP FSPSSAHFAT SLFLLLSLAG RQMSQSSLSP PSPPRSPAEE 60 EAGDKHDIHV TRATQSFPLK SIKRYTTQFL RPAPPFTSLE RFLRSKTSYG SSPLVNHSTP 120 LPSMSKSRNG TMPIRVDPGA VPPATGNNTV SSEASVIKGQ WTPEEDSLLI SLVKEHGAKK 180 WSDISKKLVG RIGKQCRERW HNHLRPDIKK DNWSDEEDLL LVRSHRKLGN RWAKIAKRIP 240 GRTENAIKNH WNATKRRQKS RRKMGKKNPS DIIINRSNIS TVLQDYILSL ETANNVITSS 300 SSSTETELIR ENTLFDSTAL IPPPSYNYHG YNGGPSSADS SPVLYQTMFE EPPSSDTMFT 360 LDPPVSPEYC MLMYPNPTSE DTVIGATSES YGLSPSVSDV NYINMSTFMS NNTDEMMMNR 420 ERQEQPLSLG MGINGENLGI IASNPTSSSS TEYKRELDLM EMMSMATYDR NSEEHYKYWC 480 NGM* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1gv2_A | 8e-45 | 156 | 258 | 2 | 104 | MYB PROTO-ONCOGENE PROTEIN |
| 1h88_C | 2e-44 | 156 | 258 | 56 | 158 | MYB PROTO-ONCOGENE PROTEIN |
| 1h89_C | 2e-44 | 156 | 258 | 56 | 158 | MYB PROTO-ONCOGENE PROTEIN |
| 1mse_C | 7e-45 | 156 | 258 | 2 | 104 | C-Myb DNA-Binding Domain |
| 1msf_C | 7e-45 | 156 | 258 | 2 | 104 | C-Myb DNA-Binding Domain |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00385 | DAP | Transfer from AT3G27785 | Download |
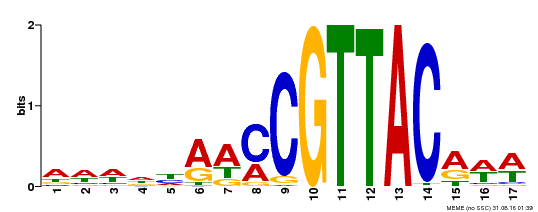
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| TrEMBL | A0A0K9NYD6 | 0.0 | A0A0K9NYD6_ZOSMR; Uncharacterized protein | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP2806 | 37 | 88 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G27785.1 | 2e-60 | myb domain protein 118 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Zosma506g00020.1 |



