 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Zosma77g00470.1 | ||||||||
| Common Name | ZOSMA_77G00470 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Alismatales; Zosteraceae; Zostera
|
||||||||
| Family | HD-ZIP | ||||||||
| Protein Properties | Length: 728aa MW: 79987.8 Da PI: 7.0809 | ||||||||
| Description | HD-ZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Homeobox | 65.1 | 1e-20 | 16 | 71 | 1 | 56 |
TT--SS--HHHHHHHHHHHHHSSS--HHHHHHHHHHCTS-HHHHHHHHHHHHHHHH CS
Homeobox 1 rrkRttftkeqleeLeelFeknrypsaeereeLAkklgLterqVkvWFqNrRakek 56
+++ +++t+ q++eLe+ F+++++p++++r +L++ lgL+ +q+k+WFqNrR+++k
Zosma77g00470.1 16 KKRYHRHTPRQIQELEATFKECPHPDEKQRMQLSRDLGLEPKQIKFWFQNRRTQMK 71
678899***********************************************998 PP
| |||||||
| 2 | START | 153.6 | 1.6e-48 | 224 | 448 | 2 | 206 |
HHHHHHHHHHHHHHHC-TT-EEEE.....EXCCTTEEEEEEESSS......SCEEEEEEEECCSCHHHHHHHHHCCCGGCT-TT-S....EEEE CS
START 2 laeeaaqelvkkalaeepgWvkss.....esengdevlqkfeeskv.....dsgealrasgvvdmvlallveellddkeqWdetla....kaet 81
la +a++el+++a+ eep+W ks+ +++n +++ + f++ + ++ea+r s + ++a+lv l+d++ +W ++ ka +
Zosma77g00470.1 224 LAASAMAELIRLAQTEEPLWIKSAltngsDILNLETYGRMFQRFNHfnnpnIRSEASRGSEMISINSATLVDLLVDTN-KWIDLFPtiisKARI 316
67899************************77777777777754333688999**************************.99888888888**** PP
EEEECTT......EEEEEEEEXXTTXX-SSX.EEEEEEEEEEE.TTS-EEEEEEEEE-TTS--.-TTSEE-EESSEEEEEEEECTCEEEEEEEE CS
START 82 levissg......galqlmvaelqalsplvp.RdfvfvRyirqlgagdwvivdvSvdseqkppesssvvRaellpSgiliepksnghskvtwve 168
lev+ +g g l+lm+ e q+lsplv+ R+f f+Ry++q ++g w+i dvSvd +++++ +++ +++lpSg+li++++ng skvtwv
Zosma77g00470.1 317 LEVLAPGipgtrsGSLVLMYEEVQVLSPLVStREFSFLRYCQQVEQGLWAITDVSVDLPREKQ-FAPSFSSKKLPSGCLIQEMPNGCSKVTWVV 409
**************************************************************9.7999999*********************** PP
-EE--SSXX.HHHHHHHHHHHHHHHHHHHHHHTXXXXXX CS
START 169 hvdlkgrlp.hwllrslvksglaegaktwvatlqrqcek 206
h+++++++p h l+r lv sg+a+ga++w+atl+r ce+
Zosma77g00470.1 410 HMEVEEKSPtHGLYRELVDSGIAFGAQRWLATLKRMCER 448
*************************************97 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF46689 | 8.77E-20 | 7 | 71 | IPR009057 | Homeodomain-like |
| Gene3D | G3DSA:1.10.10.60 | 2.2E-21 | 11 | 72 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS50071 | 17.508 | 13 | 73 | IPR001356 | Homeobox domain |
| SMART | SM00389 | 1.5E-17 | 14 | 77 | IPR001356 | Homeobox domain |
| CDD | cd00086 | 7.94E-19 | 15 | 71 | No hit | No description |
| Pfam | PF00046 | 2.6E-18 | 16 | 71 | IPR001356 | Homeobox domain |
| PROSITE pattern | PS00027 | 0 | 48 | 71 | IPR017970 | Homeobox, conserved site |
| PROSITE profile | PS50848 | 48.764 | 214 | 451 | IPR002913 | START domain |
| CDD | cd08875 | 1.28E-102 | 221 | 447 | No hit | No description |
| SuperFamily | SSF55961 | 9.06E-33 | 221 | 449 | No hit | No description |
| SMART | SM00234 | 9.2E-41 | 223 | 448 | IPR002913 | START domain |
| Pfam | PF01852 | 2.7E-41 | 224 | 448 | IPR002913 | START domain |
| Gene3D | G3DSA:3.30.530.20 | 4.0E-10 | 277 | 448 | IPR023393 | START-like domain |
| SuperFamily | SSF55961 | 2.11E-17 | 505 | 720 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009828 | Biological Process | plant-type cell wall loosening | ||||
| GO:0010091 | Biological Process | trichome branching | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0000976 | Molecular Function | transcription regulatory region sequence-specific DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0008289 | Molecular Function | lipid binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 728 aa Download sequence Send to blast |
MDFGEDPYVS DSQRRKKRYH RHTPRQIQEL EATFKECPHP DEKQRMQLSR DLGLEPKQIK 60 FWFQNRRTQM KACYLLLTQH ERADNCTLRA ENDKIRYENI AMREALKNII CPTCGGPPVS 120 EEYYFDEHKL RMENARLKEE LDRASSISSK YLGRPINQHH IPSAQPLHMM SSLDLTVGSF 180 GSNAMGPSLD LDLLPGNHSS VHHQLPFPMV LPNAAVASSF MTDLAASAMA ELIRLAQTEE 240 PLWIKSALTN GSDILNLETY GRMFQRFNHF NNPNIRSEAS RGSEMISINS ATLVDLLVDT 300 NKWIDLFPTI ISKARILEVL APGIPGTRSG SLVLMYEEVQ VLSPLVSTRE FSFLRYCQQV 360 EQGLWAITDV SVDLPREKQF APSFSSKKLP SGCLIQEMPN GCSKVTWVVH MEVEEKSPTH 420 GLYRELVDSG IAFGAQRWLA TLKRMCERIS CLMTAPDAST RELGGCGGAE GKRRMNLSQR 480 MVSNFCASLS PSTTNDSPSG TSQRWTNLTG VNDVGVRVTV HKSTDPGQPN GVVLSAATSI 540 WLPVSCNTIF NFFKDERTRS QWDVLSNGNP VQEIAHITNG SSHPGNRISL LRAFNSSQNN 600 MLILQESCSD SSGSLVVYAP VELPAINIAM SGEDPSYIPL LPSGFSILPD WRSSSGTGGG 660 GASTSSNTGS IVPGGGGSGG SLVTVAFQIL VSSLPSAKIN VESINTVNNL ISTTVQQIKS 720 ALNCTTV* |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor. {ECO:0000250}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00232 | DAP | Transfer from AT1G73360 | Download |
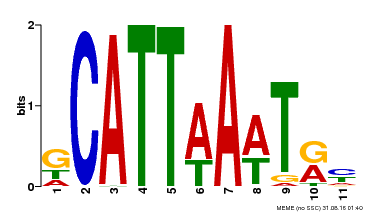
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_008806630.1 | 0.0 | homeobox-leucine zipper protein ROC8-like | ||||
| Swissprot | Q69T58 | 0.0 | ROC8_ORYSJ; Homeobox-leucine zipper protein ROC8 | ||||
| TrEMBL | A0A0K9NP46 | 0.0 | A0A0K9NP46_ZOSMR; Homeobox-leucine zipper protein ROC8 | ||||
| STRING | XP_008806630.1 | 0.0 | (Phoenix dactylifera) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP2912 | 37 | 70 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G73360.1 | 0.0 | homeodomain GLABROUS 11 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Zosma77g00470.1 |



