 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Zosma7g01530.1 | ||||||||
| Common Name | ZOSMA_7G01530 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Alismatales; Zosteraceae; Zostera
|
||||||||
| Family | MIKC_MADS | ||||||||
| Protein Properties | Length: 247aa MW: 28320 Da PI: 8.6951 | ||||||||
| Description | MIKC_MADS family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | SRF-TF | 99.2 | 1.6e-31 | 9 | 59 | 1 | 51 |
S---SHHHHHHHHHHHHHHHHHHHHHHHHHHT-EEEEEEE-TTSEEEEEE- CS
SRF-TF 1 krienksnrqvtfskRrngilKKAeELSvLCdaevaviifsstgklyeyss 51
kri+nk+nrqvtf+kRrng+lKKA+ELSvLCdaeva+iifss+gklye++s
Zosma7g01530.1 9 KRIDNKINRQVTFAKRRNGLLKKAYELSVLCDAEVALIIFSSRGKLYEFCS 59
79***********************************************96 PP
| |||||||
| 2 | K-box | 97.9 | 1.5e-32 | 80 | 173 | 7 | 100 |
K-box 7 ksleeakaeslqqelakLkkeienLqreqRhllGedLesLslkeLqqLeqqLekslkkiRskKnellleqieelqkkekelqeenkaLrkklee 100
+++++++ +s+qqe+ kLk+++e+Lqr+qR+llGedL+sLs ++L++Le+qL+ sl++iRs +++++l+q+++lqk+e++l e+nkaLr++lee
Zosma7g01530.1 80 TNVQTREIQSSQQEYLKLKARVESLQRTQRNLLGEDLGSLSSRDLENLERQLDASLRQIRSIRTQYMLDQLSDLQKQEQALCEANKALRRRLEE 173
4578999***********************************************************************************9986 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS50066 | 32.884 | 1 | 61 | IPR002100 | Transcription factor, MADS-box |
| SMART | SM00432 | 2.5E-41 | 1 | 60 | IPR002100 | Transcription factor, MADS-box |
| SuperFamily | SSF55455 | 3.27E-33 | 2 | 81 | IPR002100 | Transcription factor, MADS-box |
| CDD | cd00265 | 8.95E-45 | 2 | 75 | No hit | No description |
| PROSITE pattern | PS00350 | 0 | 3 | 57 | IPR002100 | Transcription factor, MADS-box |
| PRINTS | PR00404 | 1.8E-32 | 3 | 23 | IPR002100 | Transcription factor, MADS-box |
| Pfam | PF00319 | 9.4E-26 | 10 | 57 | IPR002100 | Transcription factor, MADS-box |
| PRINTS | PR00404 | 1.8E-32 | 23 | 38 | IPR002100 | Transcription factor, MADS-box |
| PRINTS | PR00404 | 1.8E-32 | 38 | 59 | IPR002100 | Transcription factor, MADS-box |
| Pfam | PF01486 | 8.1E-26 | 83 | 171 | IPR002487 | Transcription factor, K-box |
| PROSITE profile | PS51297 | 14.999 | 87 | 177 | IPR002487 | Transcription factor, K-box |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0001708 | Biological Process | cell fate specification | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0010093 | Biological Process | specification of floral organ identity | ||||
| GO:0048481 | Biological Process | plant ovule development | ||||
| GO:0048833 | Biological Process | specification of floral organ number | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 247 aa Download sequence Send to blast |
MGRGRVELKR IDNKINRQVT FAKRRNGLLK KAYELSVLCD AEVALIIFSS RGKLYEFCSS 60 PSMLKTLERY QKCNYVAPET NVQTREIQSS QQEYLKLKAR VESLQRTQRN LLGEDLGSLS 120 SRDLENLERQ LDASLRQIRS IRTQYMLDQL SDLQKQEQAL CEANKALRRR LEETTHPSQQ 180 QVWESEAHAM AYSRQQQSQQ QHHHQSDAFF HPLDCEPTLQ IGYHPEQITV AASGPSVGGY 240 VPGWLG* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 4ox0_A | 5e-37 | 75 | 171 | 1 | 101 | Developmental protein SEPALLATA 3 |
| 4ox0_B | 5e-37 | 75 | 171 | 1 | 101 | Developmental protein SEPALLATA 3 |
| 4ox0_C | 5e-37 | 75 | 171 | 1 | 101 | Developmental protein SEPALLATA 3 |
| 4ox0_D | 5e-37 | 75 | 171 | 1 | 101 | Developmental protein SEPALLATA 3 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor involved in flower development. {ECO:0000250|UniProtKB:Q0HA25}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00605 | ChIP-seq | Transfer from AT1G24260 | Download |
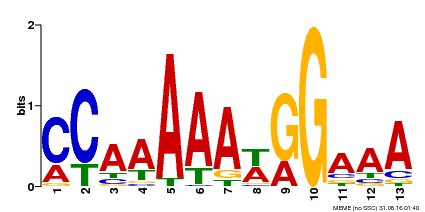
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AB474185 | 0.0 | AB474185.1 Zostera japonica ZjMADS1 mRNA for putative MADS box protein, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_008801852.1 | 1e-138 | agamous-like MADS-box protein AGL9 homolog isoform X2 | ||||
| Swissprot | Q8LLR0 | 1e-129 | MADS4_VITVI; Agamous-like MADS-box protein MADS4 | ||||
| TrEMBL | A0A0K9NPY8 | 0.0 | A0A0K9NPY8_ZOSMR; Uncharacterized protein | ||||
| STRING | GSMUA_Achr9P01370_001 | 1e-133 | (Musa acuminata) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP3483 | 34 | 69 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G24260.2 | 1e-108 | MIKC_MADS family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Zosma7g01530.1 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



