 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Zosma9g00620.1 | ||||||||
| Common Name | ZOSMA_9G00620 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Alismatales; Zosteraceae; Zostera
|
||||||||
| Family | CPP | ||||||||
| Protein Properties | Length: 789aa MW: 86293.2 Da PI: 7.3258 | ||||||||
| Description | CPP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | TCR | 50.9 | 3e-16 | 468 | 508 | 2 | 42 |
TCR 2 ekkgCnCkkskClkkYCeCfaagkkCseeCkCedCkNkeek 42
++++CnCkkskClk+YCeCfaag++C e C+C++C Nk e+
Zosma9g00620.1 468 SCRRCNCKKSKCLKLYCECFAAGVYCVEPCSCQGCLNKPEH 508
6899*********************************9876 PP
| |||||||
| 2 | TCR | 50.2 | 5e-16 | 553 | 592 | 1 | 40 |
TCR 1 kekkgCnCkkskClkkYCeCfaagkkCseeCkCedCkNke 40
++k+gCnCkks+ClkkYCeC++ g+ Cs +C+Ce+CkN+
Zosma9g00620.1 553 RHKRGCNCKKSSCLKKYCECYQGGAGCSISCRCEGCKNTF 592
589***********************************85 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SMART | SM01114 | 4.6E-16 | 467 | 508 | IPR033467 | Tesmin/TSO1-like CXC domain |
| PROSITE profile | PS51634 | 36.392 | 468 | 593 | IPR005172 | CRC domain |
| Pfam | PF03638 | 9.6E-12 | 470 | 505 | IPR005172 | CRC domain |
| SMART | SM01114 | 6.3E-17 | 553 | 594 | IPR033467 | Tesmin/TSO1-like CXC domain |
| Pfam | PF03638 | 2.4E-11 | 555 | 591 | IPR005172 | CRC domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009934 | Biological Process | regulation of meristem structural organization | ||||
| GO:0048444 | Biological Process | floral organ morphogenesis | ||||
| GO:0051302 | Biological Process | regulation of cell division | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 789 aa Download sequence Send to blast |
MDTPKRNGDG DCGKQQQSRT PMGSRVEDSP VFIYINNLSP MKPVKPVHIA QTIHSLSLPP 60 VSSIFTTPHI ATKESRFLTR SPLLSLPKPD FSDLCRSEIT NTANLPDNPD SKLSQSLNET 120 DINPTEECST RPSTQPEPFH YDCGSPYHTN SPCHDINSDT KCVMGRTSSI DFAHEDLVEK 180 IIDQSLPGFE IQSSDNDIGQ PKEVQALDWE TDTYVEEGDD LLFFNSSNLD ASVGPFHSYI 240 DHSSNVDLAT SLSQSSNDNI PTTQEENSSL RYFHVNEKID DKASENINNL FNDQGDIQQQ 300 RCMPRRCLTF DMAGVRRKSF GFREDSKTSS SLPHNSKQQT TSSTPGISRI SLHLNSAKQT 360 ASSTPGIPRI GLHLNSVASI PLDRFVQDVL PSASQHSVNM SSSIISSGNM QNKLHNSMSA 420 EKSQESDVQG LLIVQDCDTL PSDVGKSDDL SHSSPKNKKR RSETEGDSCR RCNCKKSKCL 480 KLYCECFAAG VYCVEPCSCQ GCLNKPEHKD TVLATRKQIE SRNPLAFAPK VIRSSEPVLE 540 IGEETNKTPA SARHKRGCNC KKSSCLKKYC ECYQGGAGCS ISCRCEGCKN TFGRKDGTMP 600 KGNLEADSKG GTEQQTHCSE VEKIDNHHNA TVNCNQQPIY ENLPITPFQS CRSSIQHPFT 660 SKGKPPRSSL AIGGSSSTTS KAIVKFEGPS TSYFHLLADN EIPKNLLSDG SPSSGVKVIS 720 PSKRRISPPL IEGVGGRVSP SRKGCRKFTL KSKSISSFPS LTCDSTDEYN LTKSSSPSTS 780 TAFNSQHL* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 5fd3_A | 3e-17 | 472 | 590 | 14 | 120 | Protein lin-54 homolog |
| 5fd3_B | 3e-17 | 472 | 590 | 14 | 120 | Protein lin-54 homolog |
| Search in ModeBase | ||||||
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 454 | 460 | PKNKKRR |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00624 | PBM | Transfer from PK22848.1 | Download |
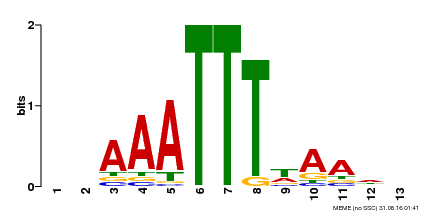
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| TrEMBL | A0A0K9NGS1 | 0.0 | A0A0K9NGS1_ZOSMR; Tesmin/TSO1-like CXC domain-containing protein | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP6147 | 34 | 47 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G22780.1 | 5e-93 | Tesmin/TSO1-like CXC domain-containing protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Zosma9g00620.1 |



