 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Zpz_sc00020.1.g00110.1.sm.mk | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; PACMAD clade; Chloridoideae; Zoysieae; Zoysiinae; Zoysia
|
||||||||
| Family | bHLH | ||||||||
| Protein Properties | Length: 473aa MW: 51379.3 Da PI: 6.146 | ||||||||
| Description | bHLH family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | HLH | 53.1 | 5.9e-17 | 288 | 334 | 4 | 55 |
HHHHHHHHHHHHHHHHHHHHHCTSCCC...TTS-STCHHHHHHHHHHHHHHH CS
HLH 4 ahnerErrRRdriNsafeeLrellPkaskapskKlsKaeiLekAveYIksLq 55
hn ErrRRdriN+++ L+el+P++ K +Ka++L +A+eY++sLq
Zpz_sc00020.1.g00110.1.sm.mk 288 VHNLSERRRRDRINEKMKALQELIPHC-----NKTDKASMLDEAIEYLRSLQ 334
6*************************8.....6******************9 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| CDD | cd00083 | 9.49E-19 | 281 | 338 | No hit | No description |
| Gene3D | G3DSA:4.10.280.10 | 6.2E-21 | 281 | 341 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SuperFamily | SSF47459 | 1.26E-20 | 282 | 344 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| PROSITE profile | PS50888 | 18.47 | 284 | 333 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Pfam | PF00010 | 2.3E-14 | 288 | 334 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SMART | SM00353 | 1.1E-18 | 290 | 339 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009704 | Biological Process | de-etiolation | ||||
| GO:0010161 | Biological Process | red light signaling pathway | ||||
| GO:0010244 | Biological Process | response to low fluence blue light stimulus by blue low-fluence system | ||||
| GO:0010600 | Biological Process | regulation of auxin biosynthetic process | ||||
| GO:0010928 | Biological Process | regulation of auxin mediated signaling pathway | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 473 aa Download sequence Send to blast |
MNHFVHDWAS SMGDAFAPLG FVPVIYGGLF LDDSGVIPFG SWLTSCESAM RFYSVEDDGL 60 MELLWCNGHV VMQSQEPRKP PRSDKEAAAV QDNEAEAWFQ CPIEDPLERD LFSELFGETP 120 ADVPSRAACK EEEDCAAVTA PQLSRPNKAC LEEIAGVSNC AVGKPDAGGD GAAATETGES 180 SMLTIGSSFC GSNHVQTTPR VVPPTPPRGE KAGARARDKR KQCDVAETEV RRTRAAGAFL 240 PRKRSITRPY RNNDTAAALQ DVEFESAVVT CEPAQKLKTA KRRRAAEVHN LSERRRRDRI 300 NEKMKALQEL IPHCNKTDKA SMLDEAIEYL RSLQLQLQVM WMGGGMATAA PVMFPAGVHQ 360 YMQRMVGPPH VAAMPRLLPF MAPPAVQSPP VGQMPVADPY AGCLAVDSLQ TPGPMHYLQG 420 VNFYQQQRPA PPPPPVVPGG SLPAATALTL PSDNTLHKEH ESSSKPETKG VNS |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 213 | 221 | ARARDKRKQ |
| 2 | 276 | 282 | LKTAKRR |
| 3 | 281 | 296 | RRRAAEVHNLSERRRR |
| 4 | 292 | 297 | ERRRRD |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00606 | ChIP-seq | Transfer from AT2G43010 | Download |
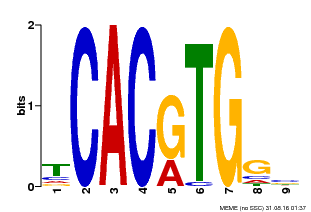
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Zpz_sc00020.1.g00110.1.sm.mk |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_025805663.1 | 1e-139 | transcription factor PHYTOCHROME INTERACTING FACTOR-LIKE 13-like isoform X3 | ||||
| TrEMBL | A0A2S3HFC1 | 1e-137 | A0A2S3HFC1_9POAL; Uncharacterized protein | ||||
| STRING | Si022196m | 1e-136 | (Setaria italica) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP3107 | 36 | 77 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G43010.2 | 8e-41 | phytochrome interacting factor 4 | ||||



