 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Zpz_sc00097.1.g00820.1.sm.mkhc | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; PACMAD clade; Chloridoideae; Zoysieae; Zoysiinae; Zoysia
|
||||||||
| Family | C2H2 | ||||||||
| Protein Properties | Length: 395aa MW: 45734.6 Da PI: 9.2577 | ||||||||
| Description | C2H2 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | zf-C2H2 | 14.7 | 8.9e-05 | 71 | 92 | 2 | 23 |
EETTTTEEESSHHHHHHHHHHT CS
zf-C2H2 2 kCpdCgksFsrksnLkrHirtH 23
+C Cg +F + +Lk+H+++H
Zpz_sc00097.1.g00820.1.sm.mkhc 71 RCLECGACFQKPAHLKQHMQRH 92
7********************9 PP
| |||||||
| 2 | zf-C2H2 | 21.3 | 7.1e-07 | 98 | 122 | 1 | 23 |
EEET..TTTEEESSHHHHHHHHHHT CS
zf-C2H2 1 ykCp..dCgksFsrksnLkrHirtH 23
+ Cp dC+ ++rk++L+rH+ tH
Zpz_sc00097.1.g00820.1.sm.mkhc 98 FNCPleDCPFIYKRKDHLNRHMLTH 122
78**********************9 PP
| |||||||
| 3 | zf-C2H2 | 12.1 | 0.00061 | 127 | 152 | 1 | 23 |
EEET..TTTEEESSHHHHHHHHHH.T CS
zf-C2H2 1 ykCp..dCgksFsrksnLkrHirt.H 23
+ C+ C+ +Fs k n++rHi+ H
Zpz_sc00097.1.g00820.1.sm.mkhc 127 FNCTvdGCDSRFSIKANMQRHIKEfH 152
679999*****************988 PP
| |||||||
| 4 | zf-C2H2 | 19.5 | 2.7e-06 | 164 | 188 | 1 | 23 |
EEET..TTTEEESSHHHHHHHHHHT CS
zf-C2H2 1 ykCp..dCgksFsrksnLkrHirtH 23
++C+ C+ksF+ +s Lk+H +H
Zpz_sc00097.1.g00820.1.sm.mkhc 164 FVCKeeGCNKSFKYSSKLKKHEESH 188
89*******************9988 PP
| |||||||
| 5 | zf-C2H2 | 22.3 | 3.4e-07 | 218 | 242 | 2 | 23 |
EET..TTTEEESSHHHHHHHHHH.T CS
zf-C2H2 2 kCp..dCgksFsrksnLkrHirt.H 23
kC+ C++sFs+ksnL++H++ H
Zpz_sc00097.1.g00820.1.sm.mkhc 218 KCSfeGCDRSFSNKSNLTKHMKAcH 242
79999*****************955 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SMART | SM00355 | 40 | 24 | 46 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE profile | PS50157 | 11.489 | 70 | 97 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 0.092 | 70 | 92 | IPR015880 | Zinc finger, C2H2-like |
| Gene3D | G3DSA:3.30.160.60 | 4.9E-6 | 71 | 96 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| PROSITE pattern | PS00028 | 0 | 72 | 92 | IPR007087 | Zinc finger, C2H2 |
| SuperFamily | SSF57667 | 1.65E-10 | 82 | 124 | No hit | No description |
| Gene3D | G3DSA:3.30.160.60 | 1.1E-7 | 97 | 124 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| PROSITE profile | PS50157 | 9.224 | 98 | 127 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 6.8E-4 | 98 | 122 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE pattern | PS00028 | 0 | 100 | 122 | IPR007087 | Zinc finger, C2H2 |
| SuperFamily | SSF57667 | 6.24E-8 | 109 | 158 | No hit | No description |
| Gene3D | G3DSA:3.30.160.60 | 8.8E-9 | 125 | 154 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| PROSITE profile | PS50157 | 10.429 | 127 | 157 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 0.02 | 127 | 152 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE pattern | PS00028 | 0 | 129 | 152 | IPR007087 | Zinc finger, C2H2 |
| Gene3D | G3DSA:3.30.160.60 | 3.9E-8 | 161 | 188 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| SuperFamily | SSF57667 | 3.6E-5 | 162 | 189 | No hit | No description |
| PROSITE profile | PS50157 | 10.72 | 164 | 188 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 0.0037 | 164 | 188 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE pattern | PS00028 | 0 | 166 | 188 | IPR007087 | Zinc finger, C2H2 |
| SuperFamily | SSF57667 | 9.82E-9 | 202 | 259 | No hit | No description |
| PROSITE profile | PS50157 | 12.695 | 217 | 247 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 7.5E-4 | 217 | 242 | IPR015880 | Zinc finger, C2H2-like |
| Gene3D | G3DSA:3.30.160.60 | 1.2E-8 | 218 | 242 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| PROSITE pattern | PS00028 | 0 | 219 | 242 | IPR007087 | Zinc finger, C2H2 |
| Gene3D | G3DSA:3.30.160.60 | 2.2E-5 | 243 | 270 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| PROSITE profile | PS50157 | 9.141 | 248 | 274 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 4.2 | 248 | 274 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE pattern | PS00028 | 0 | 250 | 274 | IPR007087 | Zinc finger, C2H2 |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0005730 | Cellular Component | nucleolus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0008097 | Molecular Function | 5S rRNA binding | ||||
| GO:0046872 | Molecular Function | metal ion binding | ||||
| GO:0080084 | Molecular Function | 5S rDNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 395 aa Download sequence Send to blast |
MCSGDEFDGG KSVGKTVYKD IRRYKCDFCD VVRSKKCLIP AHMVEHHKDE LDKSETHNSN 60 GEKIVHQVEQ RCLECGACFQ KPAHLKQHMQ RHSQERLFNC PLEDCPFIYK RKDHLNRHML 120 THQGKLFNCT VDGCDSRFSI KANMQRHIKE FHEDENAVKS NQQFVCKEEG CNKSFKYSSK 180 LKKHEESHVR LNYVEVLKKN IKRHLQAHEE APSSERMKCS FEGCDRSFSN KSNLTKHMKA 240 CHDQLKPFSC RFAGCGKAFT YKHVRDKHEQ SSAHLYVEGD FEEVDAQLHS RSRGGRKRKA 300 LTVETLTRKR VTIPSEPSAT DNAEIRSTYV EANNGPNIRS AGWEQIHRTS ACHATRPIPH 360 LGGTRRRQPS DGQHRRPQIS QHREHPDWFG PPRVA |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1tf6_A | 3e-23 | 61 | 269 | 2 | 189 | PROTEIN (TRANSCRIPTION FACTOR IIIA) |
| 1tf6_D | 3e-23 | 61 | 269 | 2 | 189 | PROTEIN (TRANSCRIPTION FACTOR IIIA) |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Essential protein (PubMed:22353599). Isoform 1 is a transcription activator the binds both 5S rDNA and 5S rRNA and stimulates the transcription of 5S rRNA gene (PubMed:12711688, PubMed:22353599). Isoform 1 regulates 5S rRNA levels during development (PubMed:22353599). {ECO:0000269|PubMed:12711688, ECO:0000269|PubMed:22353599}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00229 | DAP | Transfer from AT1G72050 | Download |
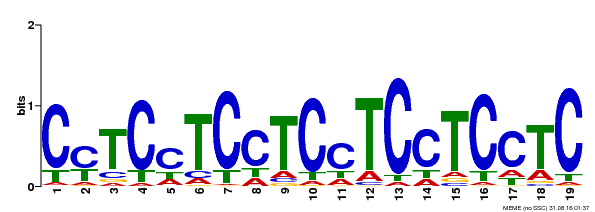
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Zpz_sc00097.1.g00820.1.sm.mkhc |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_025795605.1 | 0.0 | transcription factor IIIA-like isoform X1 | ||||
| Swissprot | Q84MZ4 | 6e-85 | TF3A_ARATH; Transcription factor IIIA | ||||
| TrEMBL | A0A2T8KTL1 | 0.0 | A0A2T8KTL1_9POAL; Uncharacterized protein | ||||
| STRING | Si020193m | 0.0 | (Setaria italica) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP2382 | 38 | 82 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G72050.2 | 2e-87 | transcription factor IIIA | ||||



