 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Zpz_sc00103.1.g00570.1.am.mk | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; PACMAD clade; Chloridoideae; Zoysieae; Zoysiinae; Zoysia
|
||||||||
| Family | WRKY | ||||||||
| Protein Properties | Length: 808aa MW: 87452.8 Da PI: 9.7172 | ||||||||
| Description | WRKY family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | WRKY | 105.6 | 2.6e-33 | 375 | 431 | 2 | 59 |
--SS-EEEEEEE--TT-SS-EEEEEE-STT---EEEEEE-SSSTTEEEEEEES--SS- CS
WRKY 2 dDgynWrKYGqKevkgsefprsYYrCtsagCpvkkkversaedpkvveitYegeHnhe 59
dDgynWrKYGqK+vkgse+prsYY+Ct+++Cp+kkkvers+ d++++ei+Y+g+Hnh
Zpz_sc00103.1.g00570.1.am.mk 375 DDGYNWRKYGQKQVKGSENPRSYYKCTFPNCPTKKKVERSQ-DGQITEIVYKGTHNHA 431
8****************************************.***************7 PP
| |||||||
| 2 | WRKY | 105.2 | 3.6e-33 | 528 | 586 | 1 | 59 |
---SS-EEEEEEE--TT-SS-EEEEEE-STT---EEEEEE-SSSTTEEEEEEES--SS- CS
WRKY 1 ldDgynWrKYGqKevkgsefprsYYrCtsagCpvkkkversaedpkvveitYegeHnhe 59
ldDgy+WrKYGqK+vkg+++prsYY+Ct+++Cpv+k+ver+++d ++v++tYeg+Hnh+
Zpz_sc00103.1.g00570.1.am.mk 528 LDDGYRWRKYGQKVVKGNPNPRSYYKCTTPSCPVRKHVERASHDLRAVITTYEGKHNHD 586
59********************************************************7 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:2.20.25.80 | 7.3E-29 | 366 | 433 | IPR003657 | WRKY domain |
| SuperFamily | SSF118290 | 2.62E-25 | 369 | 433 | IPR003657 | WRKY domain |
| PROSITE profile | PS50811 | 23.755 | 369 | 433 | IPR003657 | WRKY domain |
| SMART | SM00774 | 2.4E-36 | 374 | 432 | IPR003657 | WRKY domain |
| Pfam | PF03106 | 1.6E-25 | 375 | 430 | IPR003657 | WRKY domain |
| Gene3D | G3DSA:2.20.25.80 | 9.5E-37 | 513 | 588 | IPR003657 | WRKY domain |
| SuperFamily | SSF118290 | 4.84E-29 | 520 | 588 | IPR003657 | WRKY domain |
| PROSITE profile | PS50811 | 37.688 | 523 | 588 | IPR003657 | WRKY domain |
| SMART | SM00774 | 3.5E-38 | 528 | 587 | IPR003657 | WRKY domain |
| Pfam | PF03106 | 4.7E-25 | 529 | 586 | IPR003657 | WRKY domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009409 | Biological Process | response to cold | ||||
| GO:0009414 | Biological Process | response to water deprivation | ||||
| GO:0009651 | Biological Process | response to salt stress | ||||
| GO:0009753 | Biological Process | response to jasmonic acid | ||||
| GO:0009788 | Biological Process | negative regulation of abscisic acid-activated signaling pathway | ||||
| GO:0009938 | Biological Process | negative regulation of gibberellic acid mediated signaling pathway | ||||
| GO:0010120 | Biological Process | camalexin biosynthetic process | ||||
| GO:0010200 | Biological Process | response to chitin | ||||
| GO:0010508 | Biological Process | positive regulation of autophagy | ||||
| GO:0042742 | Biological Process | defense response to bacterium | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0050832 | Biological Process | defense response to fungus | ||||
| GO:0070370 | Biological Process | cellular heat acclimation | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| GO:0044212 | Molecular Function | transcription regulatory region DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 808 aa Download sequence Send to blast |
MPRRHRAAPR GIASPLLVVG IGFSPSFRAL ASIERRLFNA TSFDRSMSSA QLGPNNPKEL 60 GFPPPPSSRL QSPPKSHHRA SQPGQPRHRL ETARRGSIGS CDEFNFPLFR PMTTSSSGSI 120 EVPANSRPGA FSFASTSFAD MMGAGPAAAG GASRYKAMTP PSLPLSPPLV SPSSFNILGG 180 LNPADFLDSP VLLTSSFDTT PPLPCHFGAR GRGALTFSLW TYEVQDVEQS ANAFCMFQIF 240 PSPTVGAFAS QQYSWMPAQG EEQGSNNKEE QRQSYPDFSF QTPAPTSEEA VRTTTTFQTP 300 IPSASLGEET YTSQQQQPWG FQQQSGMDAG ATQAAFGSAS TFQATSSDAT TMALHVQGSG 360 AGGYGQTQSQ RRSSDDGYNW RKYGQKQVKG SENPRSYYKC TFPNCPTKKK VERSQDGQIT 420 EIVYKGTHNH AKPQNTRRNS SSAALLLQSG DASEHSFGGT PVGTPENSAS FGDDDAAGNA 480 GGDELDEDEP DSKRWRKESE AEGLSMAGNR TVREPRVVVQ TMSDIDILDD GYRWRKYGQK 540 VVKGNPNPRS YYKCTTPSCP VRKHVERASH DLRAVITTYE GKHNHDVPAA RGSAALYRPA 600 PPPPDNHYLA AAGVRPPALA YQNSGAGQQY AHGFSGQSSF GLQSATGGSF AFSGFDDLTG 660 SYMSQHQQQQ RQNDAMHASR AKEEPREDMF FQQSMMHGGP TWRDSRPRGA SGAPRWIGGR 720 WRQPARHAGL NQTAGGFVSA VSPLQTTLIR SAYRRQESGA GACGPGFALS ENTVVFVMVR 780 RQLGRRYPAR RGRGDASKIP SRGCVLPL |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1wj2_A | 6e-36 | 359 | 589 | 3 | 78 | Probable WRKY transcription factor 4 |
| 2lex_A | 6e-36 | 359 | 589 | 3 | 78 | Probable WRKY transcription factor 4 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription repressor (By similarity). Interacts specifically with the W box (5'-(T)TGAC[CT]-3'), a frequently occurring elicitor-responsive cis-acting element. Negative regulator of both gibberellic acid (GA) and abscisic acid (ABA) signaling in aleurone cells, probably by interfering with GAM1, via the specific repression of GA- and ABA-induced promoters (By similarity). {ECO:0000250|UniProtKB:Q6IEQ7, ECO:0000250|UniProtKB:Q6QHD1}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00299 | DAP | Transfer from AT2G38470 | Download |
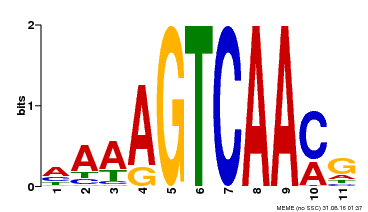
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Zpz_sc00103.1.g00570.1.am.mk |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Induced by abscisic acid (ABA) in aleurone cells, embryos, roots and leaves (PubMed:25110688). Slightly down-regulated by gibberellic acid (GA) (By similarity). Accumulates in response to jasmonic acid (MeJA) (PubMed:16919842). {ECO:0000250|UniProtKB:Q6IEQ7, ECO:0000269|PubMed:16919842, ECO:0000269|PubMed:25110688}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AY676925 | 1e-141 | AY676925.1 Oryza sativa (indica cultivar-group) transcription factor WRKY24 (WRKY24) mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_004970421.3 | 0.0 | WRKY transcription factor WRKY24 | ||||
| Swissprot | Q6B6R4 | 0.0 | WRK24_ORYSI; WRKY transcription factor WRKY24 | ||||
| TrEMBL | A0A1E5WCJ0 | 0.0 | A0A1E5WCJ0_9POAL; Putative WRKY transcription factor 33 | ||||
| STRING | Si000906m | 0.0 | (Setaria italica) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP2992 | 36 | 87 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G38470.1 | 2e-91 | WRKY DNA-binding protein 33 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



