 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Zpz_sc00885.1.g00010.1.sm.mkhc | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; PACMAD clade; Chloridoideae; Zoysieae; Zoysiinae; Zoysia
|
||||||||
| Family | WOX | ||||||||
| Protein Properties | Length: 313aa MW: 32600.1 Da PI: 8.2166 | ||||||||
| Description | WOX family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Homeobox | 47.7 | 2.7e-15 | 33 | 93 | 2 | 57 |
T--SS--HHHHHHHHHHHHH.SSS--HHHHHHHHHHC....TS-HHHHHHHHHHHHHHHHC CS
Homeobox 2 rkRttftkeqleeLeelFek.nrypsaeereeLAkkl....gLterqVkvWFqNrRakekk 57
r+R+t+++eq+ +Le+ F++ +p ++e ++ k l + +++V++WFqNrR + ++
Zpz_sc00885.1.g00010.1.sm.mkhc 33 RSRWTPKPEQILILESIFNSgMVNPPKDETVRIRKLLerfgAVGDANVFYWFQNRRSRSRR 93
89*****************99*************************************997 PP
| |||||||
| 2 | Wus_type_Homeobox | 106.9 | 1.2e-34 | 32 | 95 | 2 | 65 |
Wus_type_Homeobox 2 artRWtPtpeQikiLeelyksGlrtPnkeeiqritaeLeeyGkiedkNVfyWFQNrkaRerqkq 65
+r+RWtP+peQi iLe++++sG+++P+k+e+ ri++ Le++G+++d+NVfyWFQNr++R+r++q
Zpz_sc00885.1.g00010.1.sm.mkhc 32 VRSRWTPKPEQILILESIFNSGMVNPPKDETVRIRKLLERFGAVGDANVFYWFQNRRSRSRRRQ 95
589***********************************************************97 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:1.10.10.60 | 8.8E-9 | 28 | 100 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS50071 | 11.337 | 29 | 94 | IPR001356 | Homeobox domain |
| SuperFamily | SSF46689 | 8.13E-14 | 29 | 101 | IPR009057 | Homeodomain-like |
| SMART | SM00389 | 1.2E-6 | 31 | 98 | IPR001356 | Homeobox domain |
| Pfam | PF00046 | 5.0E-13 | 33 | 93 | IPR001356 | Homeobox domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009734 | Biological Process | auxin-activated signaling pathway | ||||
| GO:0048830 | Biological Process | adventitious root development | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 313 aa Download sequence Send to blast |
MEGSSNSPDR QSSGGSPEER GSGERQGSRE PVRSRWTPKP EQILILESIF NSGMVNPPKD 60 ETVRIRKLLE RFGAVGDANV FYWFQNRRSR SRRRQRQMQA AAAAAAAAAS SAANSHTAAA 120 SVALGVPSSG AAVHHPSAMG GSACRYEQQA SSSSSSGSTG GSSLGLFAFG AGVSGGTGGG 180 YFQTSCGASS PLGSGLIGDV DSGGGDDLFA ISRQMGFAES TVAPSSSLVP STAAQQQPQQ 240 YYSSQQLPTA TITVFINGVP MEVPRGPIDL RAMFGQDVML VHSTGTFLPV NDYGILTQSL 300 QMGESYFLVT RPT |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor which may be involved in developmental processes. {ECO:0000250}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00330 | DAP | Transfer from AT3G03660 | Download |
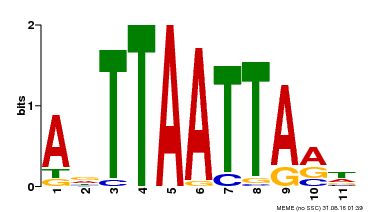
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Zpz_sc00885.1.g00010.1.sm.mkhc |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_004984518.2 | 1e-136 | WUSCHEL-related homeobox 6 | ||||
| Swissprot | A2XG77 | 1e-105 | WOX6_ORYSI; WUSCHEL-related homeobox 6 | ||||
| TrEMBL | A0A368SRG4 | 1e-135 | A0A368SRG4_SETIT; Uncharacterized protein | ||||
| STRING | Pavir.Ia03448.1.p | 1e-136 | (Panicum virgatum) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP1078 | 37 | 109 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G17810.2 | 3e-32 | WUSCHEL related homeobox 12 | ||||



