 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Zpz_sc01002.1.g00140.1.am.mk | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; PACMAD clade; Chloridoideae; Zoysieae; Zoysiinae; Zoysia
|
||||||||
| Family | ERF | ||||||||
| Protein Properties | Length: 391aa MW: 42058.1 Da PI: 9.2854 | ||||||||
| Description | ERF family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | AP2 | 57.2 | 4.1e-18 | 47 | 96 | 2 | 55 |
AP2 2 gykGVrwdkkrgrWvAeIrdpsengkrkrfslgkfgtaeeAakaaiaarkkleg 55
++ GVr+++ +grWvAeI+d + ++ r +lg+f taeeAa+a+++a+ l+g
Zpz_sc01002.1.g00140.1.am.mk 47 KFVGVRQRP-SGRWVAEIKDTT---QKIRMWLGTFETAEEAARAYDEAACLLRG 96
799******.7*********32...35**********************99998 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF00847 | 9.9E-13 | 47 | 96 | IPR001471 | AP2/ERF domain |
| Gene3D | G3DSA:3.30.730.10 | 1.4E-27 | 47 | 104 | IPR001471 | AP2/ERF domain |
| CDD | cd00018 | 3.54E-29 | 47 | 103 | No hit | No description |
| PROSITE profile | PS51032 | 19.836 | 47 | 104 | IPR001471 | AP2/ERF domain |
| SMART | SM00380 | 1.2E-33 | 47 | 110 | IPR001471 | AP2/ERF domain |
| SuperFamily | SSF54171 | 6.54E-19 | 47 | 104 | IPR016177 | DNA-binding domain |
| PRINTS | PR00367 | 1.7E-7 | 48 | 59 | IPR001471 | AP2/ERF domain |
| PRINTS | PR00367 | 1.7E-7 | 70 | 86 | IPR001471 | AP2/ERF domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0000302 | Biological Process | response to reactive oxygen species | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009723 | Biological Process | response to ethylene | ||||
| GO:0035865 | Biological Process | cellular response to potassium ion | ||||
| GO:0048528 | Biological Process | post-embryonic root development | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 391 aa Download sequence Send to blast |
MELQFQKPQS HAQQQQCQYQ AVSVVAKETA GKLSRARSKC GGGGGGKFVG VRQRPSGRWV 60 AEIKDTTQKI RMWLGTFETA EEAARAYDEA ACLLRGANTR TNFAAAGGAG SAPPVSPLAS 120 RIRALLHHKK FKKTVARQPD VAFSPPAAAA YRHGGGTGSV AATSTSASTV TTTTSSVSPS 180 SSPSSTTIKF AMTSGGIRTP ILPVPSHHTF TEESACYRPY LLTTGSDEFQ HQYEQSWTLN 240 TSSLPPTDGR DNACSRMPEL EKISPEKDGP ASPPHGEDRV QGGQDFFDAG NDASDSLVRT 300 FWVRFDLLDC SCRGANSVQK CMRKGEQQSR SPAVIAADQL AIAMATAGVS SAAERQRPSC 360 KEEVVQLSRL LGDSPSILAL QRFRWFPFYF S |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 5wx9_A | 1e-13 | 46 | 103 | 13 | 71 | Ethylene-responsive transcription factor ERF096 |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00521 | DAP | Transfer from AT5G19790 | Download |
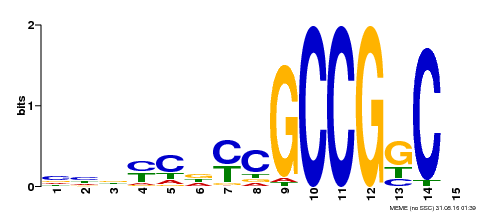
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Zpz_sc01002.1.g00140.1.am.mk |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | - | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AP004273 | 2e-57 | AP004273.2 Oryza sativa Japonica Group genomic DNA, chromosome 7, PAC clone:P0431A02. | |||
| GenBank | AP014963 | 2e-57 | AP014963.1 Oryza sativa Japonica Group DNA, chromosome 7, cultivar: Nipponbare, complete sequence. | |||
| GenBank | CP012615 | 2e-57 | CP012615.1 Oryza sativa Indica Group cultivar RP Bio-226 chromosome 7 sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_002459520.2 | 2e-78 | ethylene-responsive transcription factor RAP2-11 isoform X1 | ||||
| TrEMBL | A0A1B6Q9T2 | 4e-77 | A0A1B6Q9T2_SORBI; Uncharacterized protein | ||||
| STRING | Sb02g006023.1 | 8e-78 | (Sorghum bicolor) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP3900 | 32 | 73 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G19790.1 | 2e-22 | related to AP2 11 | ||||



