 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Zpz_sc01213.1.g00140.1.am.mk | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; PACMAD clade; Chloridoideae; Zoysieae; Zoysiinae; Zoysia
|
||||||||
| Family | bHLH | ||||||||
| Protein Properties | Length: 607aa MW: 64183.7 Da PI: 10.3901 | ||||||||
| Description | bHLH family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | HLH | 50 | 5.4e-16 | 403 | 448 | 5 | 55 |
HHHHHHHHHHHHHHHHHHHHCTSCCC...TTS-STCHHHHHHHHHHHHHHH CS
HLH 5 hnerErrRRdriNsafeeLrellPkaskapskKlsKaeiLekAveYIksLq 55
hne Er+RRdriN+++ +L++l+P++ K +Ka++L ++++Y+k+Lq
Zpz_sc01213.1.g00140.1.am.mk 403 HNESERKRRDRINQKMKTLQKLVPNS-----SKTDKASMLDEVIDYLKQLQ 448
*************************8.....7******************9 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:4.10.280.10 | 1.3E-17 | 396 | 462 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| PROSITE profile | PS50888 | 18.025 | 398 | 447 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SuperFamily | SSF47459 | 1.09E-19 | 401 | 471 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Pfam | PF00010 | 1.5E-13 | 403 | 448 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| CDD | cd00083 | 2.77E-17 | 403 | 452 | No hit | No description |
| SMART | SM00353 | 1.2E-18 | 404 | 453 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009567 | Biological Process | double fertilization forming a zygote and endosperm | ||||
| GO:0009506 | Cellular Component | plasmodesma | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 607 aa Download sequence Send to blast |
MWKRRRLRAR LRLTKPRPLA ASSSSVPSLS LPPPPPIRTP GHAHHRSTVR AAWLPPHTTP 60 RTHRRQCFKY LLLRAIRSPR PPHRASTRAL AARSNRALPA SLTAAPVSSR LTSSHATGRG 120 GGAAEMNQCV PSWDLDDAAA GGLNTASAAT DGVHHRAGVD GAGYYDVAEL TWEKGNISSH 180 GLLNRPAPTK CPPAASSTPL QALGGGGEPA GGGDRETLEA VVGEAAARSH FPAQPSLPTT 240 PWLGVGVPLP AEALVPCATR DDQAAEEEAR RKRARGVGED GHGLVCASQG STAPGRRGES 300 ALPELDAACG IGDDDVCGFT TTTNNSTSLD RDDNKGSPDT ENTSISGGAS DSRCFSLRSQ 360 ANCLLITLPR DGLCDEGENV VINGEPAMRS SVSTKRSRAA AIHNESERKR RDRINQKMKT 420 LQKLVPNSSK TDKASMLDEV IDYLKQLQAQ VQVMSRMSSM MMPMAMPQLQ MSVMAQMAQM 480 AQMAQMAQGM MNMGSLSQPG YVPPPMMHPP PPPFVPLPWD AGAAGASSGA GAVXXXXAAA 540 PPLRTPSRPS SRANKRSRMA RYVMQPQIVS ALSSSRAMRG TDEVKFRAAA AATGLHGGVQ 600 QDVGDVA |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 2 | 6 | KRRRL |
| 2 | 3 | 10 | RRRLRARL |
| 3 | 406 | 411 | ERKRRD |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00034 | PBM | Transfer from AT4G00050 | Download |
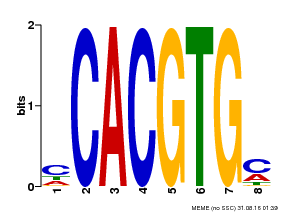
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Zpz_sc01213.1.g00140.1.am.mk |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | - | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AK361420 | 1e-111 | AK361420.1 Hordeum vulgare subsp. vulgare mRNA for predicted protein, complete cds, clone: NIASHv1140E09. | |||
| GenBank | AK367814 | 1e-111 | AK367814.1 Hordeum vulgare subsp. vulgare mRNA for predicted protein, complete cds, clone: NIASHv2062O04. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_004983681.1 | 1e-140 | transcription factor UNE10 isoform X1 | ||||
| TrEMBL | K4A8X8 | 1e-138 | K4A8X8_SETIT; Uncharacterized protein | ||||
| STRING | Si035334m | 1e-139 | (Setaria italica) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP7474 | 31 | 40 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G00050.1 | 6e-33 | bHLH family protein | ||||



