 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Zpz_sc01473.1.g00050.1.am.mkhc | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; PACMAD clade; Chloridoideae; Zoysieae; Zoysiinae; Zoysia
|
||||||||
| Family | bZIP | ||||||||
| Protein Properties | Length: 287aa MW: 31110.1 Da PI: 6.2589 | ||||||||
| Description | bZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | bZIP_1 | 69.2 | 6.5e-22 | 190 | 252 | 1 | 63 |
XXXXCHHHCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHH CS
bZIP_1 1 ekelkrerrkqkNReAArrsRqRKkaeieeLeekvkeLeaeNkaLkkeleelkkevaklksev 63
e+elkr+rrkq+NRe+ArrsR+RK+ae+eeL++ +++L++eN++L++e+++++ke+++l s++
Zpz_sc01473.1.g00050.1.am.mkhc 190 ERELKRQRRKQSNRESARRSRLRKQAECEELAQHAEVLKQENTSLRDEVNRIRKEYEELLSKN 252
89**********************************************************998 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF07777 | 1.5E-11 | 1 | 93 | IPR012900 | G-box binding protein, multifunctional mosaic region |
| Pfam | PF16596 | 2.9E-14 | 96 | 165 | No hit | No description |
| Gene3D | G3DSA:1.20.5.170 | 2.1E-18 | 184 | 248 | No hit | No description |
| SMART | SM00338 | 7.9E-20 | 190 | 254 | IPR004827 | Basic-leucine zipper domain |
| Pfam | PF00170 | 2.3E-20 | 190 | 252 | IPR004827 | Basic-leucine zipper domain |
| PROSITE profile | PS50217 | 12.955 | 192 | 255 | IPR004827 | Basic-leucine zipper domain |
| SuperFamily | SSF57959 | 7.5E-11 | 193 | 249 | No hit | No description |
| CDD | cd14702 | 3.48E-23 | 195 | 245 | No hit | No description |
| PROSITE pattern | PS00036 | 0 | 197 | 212 | IPR004827 | Basic-leucine zipper domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 287 aa Download sequence Send to blast |
MGSSGTDTPA KSSKTSATQE QQPPASLNVA TPAVYPDWSR FQGYPPIPPH GFFPSPVPSS 60 AQANDTALWD PTTSICHGAH PFTPYAITSP NGNADASDSQ KGSGQEQDGD VRSSQNGVSR 120 SPSEGKLNQT MAIMPMPSSG PVPGPTTNLN IGMDYWGANT ASSTPAIHDR GTPTAVPGAV 180 VPAEQWIQDE RELKRQRRKQ SNRESARRSR LRKQAECEEL AQHAEVLKQE NTSLRDEVNR 240 IRKEYEELLS KNNLLKENLG GKLDETDETG LDNKLQHSGG EIRNKGN |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 206 | 212 | RRSRLRK |
| 2 | 206 | 213 | RRSRLRKQ |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor that may be involved in responses to fungal pathogen infection and abiotic stresses. {ECO:0000269|Ref.1}. | |||||
| UniProt | Probable transcription factor that may be involved in responses to fungal pathogen infection and abiotic stresses. {ECO:0000269|Ref.2}. | |||||
| UniProt | Probable transcription factor that may be involved in responses to fungal pathogen infection and abiotic stresses. {ECO:0000269|Ref.2}. | |||||
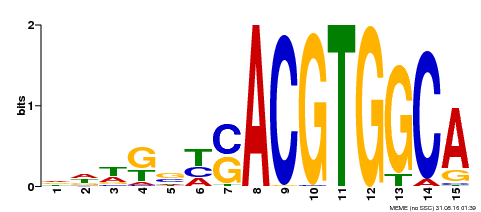
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00291 | DAP | Transfer from AT2G35530 | Download |
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Zpz_sc01473.1.g00050.1.am.mkhc |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Induced during incompatible interaction with the fungal pathogen Puccinia striiformis (Ref.1). Induced by abscisic acid (ABA), ethylene, cold stress, salt stress and wounding (Ref.1). {ECO:0000269|Ref.1}. | |||||
| UniProt | INDUCTION: Induced during incompatible interaction with the fungal pathogen Puccinia striiformis (Ref.2). Induced by abscisic acid (ABA), ethylene, cold stress, salt stress and wounding (Ref.2). {ECO:0000269|Ref.2}. | |||||
| UniProt | INDUCTION: Induced during incompatible interaction with the fungal pathogen Puccinia striiformis (Ref.2). Induced by abscisic acid (ABA), ethylene, cold stress, salt stress and wounding (Ref.2). {ECO:0000269|Ref.2}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | - | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | NP_001131383.1 | 1e-107 | uncharacterized protein LOC100192709 | ||||
| Refseq | XP_008672076.1 | 1e-107 | putative bZIP transcription factor superfamily protein isoform X1 | ||||
| Refseq | XP_008672077.1 | 1e-107 | putative bZIP transcription factor superfamily protein isoform X1 | ||||
| Swissprot | A0A3B6KF13 | 4e-97 | BZP1A_WHEAT; bZIP transcription factor 1-A | ||||
| Swissprot | A0A3B6MPP5 | 4e-97 | BZP1D_WHEAT; bZIP transcription factor 1-D | ||||
| Swissprot | B6E107 | 4e-97 | BZP1B_WHEAT; bZIP transcription factor 1-B | ||||
| TrEMBL | A0A3L6SVN0 | 1e-107 | A0A3L6SVN0_PANMI; Putative bZIP transcription factor superfamily protein | ||||
| STRING | GRMZM2G074373_P01 | 1e-107 | (Zea mays) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP2147 | 37 | 96 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G35530.1 | 6e-40 | basic region/leucine zipper transcription factor 16 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



