 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Zpz_sc02100.1.g00110.1.am.mk | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; PACMAD clade; Chloridoideae; Zoysieae; Zoysiinae; Zoysia
|
||||||||
| Family | NAC | ||||||||
| Protein Properties | Length: 524aa MW: 56924.9 Da PI: 5.6202 | ||||||||
| Description | NAC family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | NAM | 176 | 1.1e-54 | 111 | 240 | 1 | 129 |
NAM 1 lppGfrFhPtdeelvveyLkkkvegkkleleevikevdiykvePwdLpk...kvkaeekewyfFskrdkkyatgkrknrat 78
+ppGfrFhPtdeelv +yL+kkv+++k++l +vi+++d+y++ePwdL++ + e++ewyfFs +d+ky+tg+r+nrat
Zpz_sc02100.1.g00110.1.am.mk 111 VPPGFRFHPTDEELVGYYLRKKVASQKIDL-DVIRDIDLYRIEPWDLQEhcgIGYDEQNEWYFFSYKDRKYPTGTRTNRAT 190
69****************************.9***************953432234677********************** PP
NAM 79 ksgyWkatgkdkevlskkgelvglkktLvfykgrapkgektdWvmheyrle 129
+g+Wkatg+dk+v++ k++l+g++ktLvfykgrap+g+ktdW+mheyrle
Zpz_sc02100.1.g00110.1.am.mk 191 MAGFWKATGRDKAVHD-KSRLIGMRKTLVFYKGRAPNGQKTDWIMHEYRLE 240
****************.999*****************************85 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF101941 | 6.54E-57 | 109 | 249 | IPR003441 | NAC domain |
| PROSITE profile | PS51005 | 56.488 | 111 | 298 | IPR003441 | NAC domain |
| Pfam | PF02365 | 3.2E-28 | 112 | 239 | IPR003441 | NAC domain |
| SuperFamily | SSF101941 | 6.54E-57 | 286 | 298 | IPR003441 | NAC domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0010200 | Biological Process | response to chitin | ||||
| GO:0048759 | Biological Process | xylem vessel member cell differentiation | ||||
| GO:1901348 | Biological Process | positive regulation of secondary cell wall biogenesis | ||||
| GO:1990110 | Biological Process | callus formation | ||||
| GO:0044212 | Molecular Function | transcription regulatory region DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 524 aa Download sequence Send to blast |
MVSFCAMEFL DQATERSEAV QATETHLLLV LPGFSLAILP RPVHARHAFV APLDGGDGGP 60 STLSFAGLYL YPHLSLPPAE ALPGSGVAES DAHTYSGAAP GDHMDSIESC VPPGFRFHPT 120 DEELVGYYLR KKVASQKIDL DVIRDIDLYR IEPWDLQEHC GIGYDEQNEW YFFSYKDRKY 180 PTGTRTNRAT MAGFWKATGR DKAVHDKSRL IGMRKTLVFY KGRAPNGQKT DWIMHEYRLE 240 TDENAPPQAR SLAPLLVIYL LVSGSVNVLA RAISRFTPNK LVVVLQEEGW VVCRAFKKRT 300 AYHARSMALA WDSSFAYRDP NVMVGATAAE AAAFVDPNEA YAQIRRQSSK NARFKQEAEL 360 DGAAAFLQYS SHLVELPQLE SPSAPLAPNT SHASTEEGGS GGGRRSKKAR ADKVATDWRA 420 LDKFVASQLS PGEGGDIEAT ASAAAAGEGS QLDHGCEEDM AALLFLNSDG REEVERWTGL 480 LGPASGDGDL GFSPVATCDP SLLTNFVVGP SPAGGAADLR RGRD |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 3ulx_A | 2e-46 | 108 | 239 | 12 | 140 | Stress-induced transcription factor NAC1 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription activator that binds to the secondary wall NAC binding element (SNBE), 5'-(T/A)NN(C/T)(T/C/G)TNNNNNNNA(A/C)GN(A/C/T)(A/T)-3', in the promoter of target genes (By similarity). Involved in xylem formation by promoting the expression of secondary wall-associated transcription factors and of genes involved in secondary wall biosynthesis and programmed cell death, genes driven by the secondary wall NAC binding element (SNBE). Triggers thickening of secondary walls (PubMed:25148240). {ECO:0000250|UniProtKB:Q9LVA1, ECO:0000269|PubMed:25148240}. | |||||
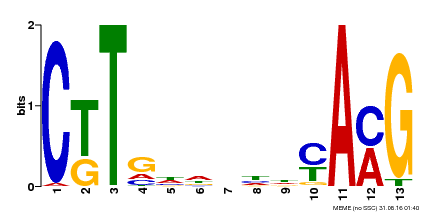
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00266 | DAP | Transfer from AT2G18060 | Download |
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Zpz_sc02100.1.g00110.1.am.mk |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Induced by chitin (e.g. chitooctaose) (PubMed:17722694). Accumulates in plants exposed to callus induction medium (CIM) (PubMed:17581762). {ECO:0000269|PubMed:17581762, ECO:0000269|PubMed:17722694}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | - | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_022678630.1 | 0.0 | NAC domain-containing protein 37 isoform X1 | ||||
| Swissprot | Q9SL41 | 1e-116 | NAC37_ARATH; NAC domain-containing protein 37 | ||||
| TrEMBL | A0A368SWG2 | 0.0 | A0A368SWG2_SETIT; Uncharacterized protein | ||||
| STRING | Si036363m | 0.0 | (Setaria italica) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP3625 | 37 | 68 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G36160.1 | 1e-111 | NAC domain containing protein 76 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



