 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Zpz_sc02195.1.g00040.1.sm.mkhc | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; PACMAD clade; Chloridoideae; Zoysieae; Zoysiinae; Zoysia
|
||||||||
| Family | MYB_related | ||||||||
| Protein Properties | Length: 305aa MW: 32459.6 Da PI: 10.4301 | ||||||||
| Description | MYB_related family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 48.2 | 2.5e-15 | 121 | 165 | 3 | 47 |
SS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHH CS
Myb_DNA-binding 3 rWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqky 47
+WT++E+ +++ + ++lG+g+W+ I+r++ ++Rt+ q+ s+ qky
Zpz_sc02195.1.g00040.1.sm.mkhc 121 PWTEDEHRRFLAGLEKLGKGDWRGISRHFVTTRTPTQVASHAQKY 165
8*******************************************9 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS50158 | 8.68 | 3 | 18 | IPR001878 | Zinc finger, CCHC-type |
| PROSITE profile | PS51294 | 20.171 | 114 | 170 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 3.14E-18 | 115 | 171 | IPR009057 | Homeodomain-like |
| TIGRFAMs | TIGR01557 | 8.6E-19 | 117 | 169 | IPR006447 | Myb domain, plants |
| SMART | SM00717 | 1.5E-11 | 118 | 168 | IPR001005 | SANT/Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 2.3E-11 | 118 | 164 | IPR009057 | Homeodomain-like |
| CDD | cd00167 | 6.05E-10 | 121 | 166 | No hit | No description |
| Pfam | PF00249 | 2.5E-12 | 121 | 165 | IPR001005 | SANT/Myb domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0008270 | Molecular Function | zinc ion binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 305 aa Download sequence Send to blast |
MARKCSSCGN NGHNSRTCSG GYRSMESTGV CGSSGGVRLF GVQLHVGSSP TPMKKCFSME 60 CLSSSAPAYY AAALAGACSS SPSASSSSSL VSVDETAEKM TNGYLSDGLM GRAQERKKGV 120 PWTEDEHRRF LAGLEKLGKG DWRGISRHFV TTRTPTQVAS HAQKYFLRQS SLTQKKRRSS 180 LFDVVENAEG PTRVNGRLKL RDETSSMSET EFPALSLGVS RRPKPEHALP PGLALAPRFS 240 ASTSSSGSAS PNLIAPRHPS LTTPKPAPVS LQAPDLELKI STARPTDQAA GPPPRTPFFG 300 TIRVT |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00565 | DAP | Transfer from AT5G56840 | Download |
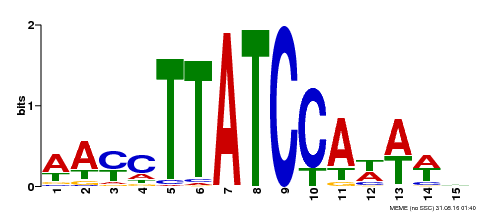
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Zpz_sc02195.1.g00040.1.sm.mkhc |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_015625837.1 | 1e-113 | transcription factor MYBS3-like | ||||
| TrEMBL | A0A0A9ESZ5 | 1e-144 | A0A0A9ESZ5_ARUDO; Uncharacterized protein | ||||
| STRING | ORUFI01G06450.1 | 1e-117 | (Oryza rufipogon) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP4231 | 38 | 73 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G56840.1 | 2e-45 | MYB_related family protein | ||||



