 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Zpz_sc04068.1.g00040.1.sm.mkhc | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; PACMAD clade; Chloridoideae; Zoysieae; Zoysiinae; Zoysia
|
||||||||
| Family | HD-ZIP | ||||||||
| Protein Properties | Length: 699aa MW: 75746.4 Da PI: 6.4156 | ||||||||
| Description | HD-ZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Homeobox | 51.3 | 2e-16 | 10 | 49 | 17 | 56 |
HHHHHSSS--HHHHHHHHHHCTS-HHHHHHHHHHHHHHHH CS
Homeobox 17 elFeknrypsaeereeLAkklgLterqVkvWFqNrRakek 56
+lF+++++p++++r eL+k+lgL+ rqVk+WFqNrR+++k
Zpz_sc04068.1.g00040.1.sm.mkhc 10 RLFKECPHPDEKQRGELSKRLGLDPRQVKFWFQNRRTQMK 49
79***********************************999 PP
| |||||||
| 2 | START | 190.9 | 6.3e-60 | 209 | 441 | 1 | 206 |
HHHHHHHHHHHHHHHHC-TT-EEEE......EXCCTTEEEEEEESSS......SCEEEEEEEECCSC.HHHHHHHHHCC CS
START 1 elaeeaaqelvkkalaeepgWvkss......esengdevlqkfeeskv.....dsgealrasgvvdm.vlallveelld 67
ela +a++elvk+a+++ep+W + e +n +e+ ++f++ + + +ea+r+sg+v+ ++a lve+l+d
Zpz_sc04068.1.g00040.1.sm.mkhc 209 ELAISAMDELVKMAQVDEPLWLTNMpgapnkETLNFEEYFRSFVPCLGmkpvgFVSEASRESGLVIIdNSAALVETLMD 287
57889*****************999899999999999999999886669999***********997615669******9 PP
CGGCT-TT-S....EEEEEEEECTT......EEEEEEEEXXTTXX-SSX.EEEEEEEEEEE.TTS-EEEEEEEEE.... CS
START 68 dkeqWdetla....kaetlevissg......galqlmvaelqalsplvp.RdfvfvRyirqlgagdwvivdvSvd.... 131
++ +W+ ++ ka++le +s+g g+l lm+aelq+lsplvp R+++f+R+++ql +g w++vdvSvd
Zpz_sc04068.1.g00040.1.sm.mkhc 288 KR-RWSDMFScmiaKATILEEVSTGiaggrnGVLLLMKAELQVLSPLVPiREVTFLRFSKQLAEGAWAVVDVSVDgllr 365
99.***********************************************************************84422 PP
.-TTS--....-TTSEE-EESSEEEEEEEECTCEEEEEEEE-EE--SSXXHHHHHHHHHHHHHHHHHHHHHHTXXXXXX CS
START 132 .seqkppe...sssvvRaellpSgiliepksnghskvtwvehvdlkgrlphwllrslvksglaegaktwvatlqrqcek 206
+++ +++ v++++lpSg+++++++ng++kvtwveh++++++++h+l+r+l++sgla+ga++w+atlqrqce+
Zpz_sc04068.1.g00040.1.sm.mkhc 366 dQNS---AttsTAGNVKCRRLPSGCVMQDTPNGYCKVTWVEHTEYDEASVHQLYRPLLRSGLAFGARRWLATLQRQCEC 441
1233...24569*****************************************************************96 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SMART | SM00389 | 8.4E-9 | 5 | 55 | IPR001356 | Homeobox domain |
| PROSITE profile | PS50071 | 14.544 | 8 | 51 | IPR001356 | Homeobox domain |
| SuperFamily | SSF46689 | 1.17E-13 | 9 | 55 | IPR009057 | Homeodomain-like |
| CDD | cd00086 | 7.77E-14 | 9 | 51 | No hit | No description |
| Gene3D | G3DSA:1.10.10.60 | 5.4E-15 | 10 | 58 | IPR009057 | Homeodomain-like |
| Pfam | PF00046 | 7.9E-14 | 10 | 49 | IPR001356 | Homeobox domain |
| PROSITE pattern | PS00027 | 0 | 26 | 49 | IPR017970 | Homeobox, conserved site |
| PROSITE profile | PS50848 | 46.093 | 200 | 444 | IPR002913 | START domain |
| SuperFamily | SSF55961 | 9.42E-30 | 203 | 441 | No hit | No description |
| CDD | cd08875 | 4.43E-116 | 204 | 440 | No hit | No description |
| SMART | SM00234 | 3.5E-46 | 209 | 441 | IPR002913 | START domain |
| Pfam | PF01852 | 1.6E-52 | 210 | 441 | IPR002913 | START domain |
| SuperFamily | SSF55961 | 7.85E-22 | 461 | 680 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0048497 | Biological Process | maintenance of floral organ identity | ||||
| GO:0008289 | Molecular Function | lipid binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 699 aa Download sequence Send to blast |
MDSCPCCVRR LFKECPHPDE KQRGELSKRL GLDPRQVKFW FQNRRTQMKT QLERHENALL 60 KQENDKLRAE NMTIREAMRS PMCGGCGSPA MLGEVSLEEQ HLRIENARLK DELNRVYSLA 120 TKFLGKPLSF MSGYQMQPHL ALPMPSSSLE LAVGGLGGMG SMPSAAMPGS MGEFTGGVSS 180 PMGTVITPAR AAGAAPPPMV GVDRSILLEL AISAMDELVK MAQVDEPLWL TNMPGAPNKE 240 TLNFEEYFRS FVPCLGMKPV GFVSEASRES GLVIIDNSAA LVETLMDKRR WSDMFSCMIA 300 KATILEEVST GIAGGRNGVL LLMKAELQVL SPLVPIREVT FLRFSKQLAE GAWAVVDVSV 360 DGLLRDQNSA TTSTAGNVKC RRLPSGCVMQ DTPNGYCKVT WVEHTEYDEA SVHQLYRPLL 420 RSGLAFGARR WLATLQRQCE CLAILMSPDT VSANDASVIT QEGKRSMLKL ARRMMENFCA 480 GVSASSAREW SKLDGAAGSI GEDVRVMARK SVNEPGEPPG VVLSAATSVW VPVAPEKLFN 540 FLRNEQLRAE WDILSNGGPM QEMANIAKGQ GPGNSVSLLR ASAMSANQSS MLILQETCTD 600 ASGSMVVYAP VDIPAMQLVM NGGDSTYVAL LPSGFAILPD APGIDSAEHK TGGSLLTVAF 660 QILVNSQPTA KLTVESVETV NNLISCTIKK IKNALQCDT |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor. {ECO:0000250}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00418 | DAP | Transfer from AT3G61150 | Download |
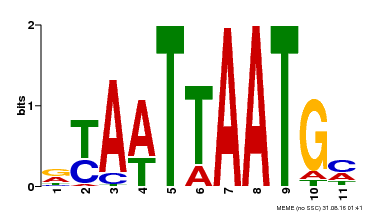
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Zpz_sc04068.1.g00040.1.sm.mkhc |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_002457484.1 | 0.0 | homeobox-leucine zipper protein ROC5 | ||||
| Swissprot | Q6EPF0 | 0.0 | ROC5_ORYSJ; Homeobox-leucine zipper protein ROC5 | ||||
| TrEMBL | C5XEA6 | 0.0 | C5XEA6_SORBI; Uncharacterized protein | ||||
| STRING | Sb03g008090.1 | 0.0 | (Sorghum bicolor) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP1203 | 37 | 116 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G61150.1 | 0.0 | homeodomain GLABROUS 1 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



