 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | augustus_masked-scaffold00494-abinit-gene-0.12-mRNA-1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Fagales; Fagaceae; Castanea
|
||||||||
| Family | ERF | ||||||||
| Protein Properties | Length: 303aa MW: 33276.5 Da PI: 7.218 | ||||||||
| Description | ERF family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | AP2 | 65.4 | 1.2e-20 | 175 | 224 | 2 | 55 |
AP2 2 gykGVrwdkkrgrWvAeIrdpsengkr.krfslgkfgtaeeAakaaiaarkkleg 55
y+GVr+++ +g+W+AeIrdp+ + +r++lg+f taeeAa+a+++a+ +++g
augustus_masked-scaffold00494-abinit-gene-0.12-mRNA-1 175 NYRGVRQRP-WGKWAAEIRDPR----KaTRVWLGTFTTAEEAARAYDRAAIEFRG 224
59*******.**********94....36************************998 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51032 | 25.277 | 175 | 232 | IPR001471 | AP2/ERF domain |
| SMART | SM00380 | 2.7E-40 | 175 | 238 | IPR001471 | AP2/ERF domain |
| Gene3D | G3DSA:3.30.730.10 | 3.4E-33 | 175 | 232 | IPR001471 | AP2/ERF domain |
| Pfam | PF00847 | 2.8E-14 | 175 | 224 | IPR001471 | AP2/ERF domain |
| SuperFamily | SSF54171 | 2.62E-23 | 175 | 234 | IPR016177 | DNA-binding domain |
| CDD | cd00018 | 5.12E-36 | 175 | 232 | No hit | No description |
| PRINTS | PR00367 | 4.6E-12 | 176 | 187 | IPR001471 | AP2/ERF domain |
| PRINTS | PR00367 | 4.6E-12 | 198 | 214 | IPR001471 | AP2/ERF domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0001944 | Biological Process | vasculature development | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0010200 | Biological Process | response to chitin | ||||
| GO:0050832 | Biological Process | defense response to fungus | ||||
| GO:0051301 | Biological Process | cell division | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 303 aa Download sequence Send to blast |
MYNPSKRTKI NQNPISQHSS LTPPPMPPLQ HHRLSTEDEL SVMVAALTNV VSGSSSTSAA 60 ATATANQQFY GYPWLPSPPT FDGAAAASTS SPTSTGTDCT DTFLPPSSEL DTCHVCNIDG 120 CLGCNFFPPP PTQQYTTTDY HHRRQHQQEK KIAVSSGSGS SKKQTQQKQQ RVKKNYRGVR 180 QRPWGKWAAE IRDPRKATRV WLGTFTTAEE AARAYDRAAI EFRGPRAKLN FPFSDYTGAN 240 SSNSSDLRTT AENSAEIEVT NLIGNGNQDN NFVDEHWEEW MMTLMNGGDS SDSAGNTSNA 300 LSY |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1gcc_A | 4e-24 | 174 | 233 | 1 | 61 | ETHYLENE RESPONSIVE ELEMENT BINDING FACTOR 1 |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00469 | DAP | Transfer from AT4G34410 | Download |
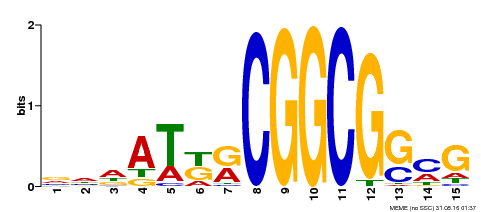
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_023896297.1 | 1e-159 | ethylene-responsive transcription factor ERF109 | ||||
| TrEMBL | A0A2N9HVK1 | 1e-92 | A0A2N9HVK1_FAGSY; Uncharacterized protein | ||||
| STRING | EOX98256 | 1e-68 | (Theobroma cacao) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF2962 | 30 | 72 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G34410.1 | 2e-34 | redox responsive transcription factor 1 | ||||



