 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | augustus_masked-scaffold00762-abinit-gene-0.10-mRNA-1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Fagales; Fagaceae; Castanea
|
||||||||
| Family | GATA | ||||||||
| Protein Properties | Length: 335aa MW: 36473.1 Da PI: 8.6758 | ||||||||
| Description | GATA family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | GATA | 58.4 | 9.8e-19 | 248 | 281 | 1 | 34 |
GATA 1 CsnCgttkTplWRrgpdgnktLCnaCGlyyrkkg 34
C +C +tkTp+WR+gp g+ktLCnaCG++yr+ +
augustus_masked-scaffold00762-abinit-gene-0.10-mRNA-1 248 CMHCEVTKTPQWREGPMGPKTLCNACGVRYRSGR 281
99*****************************987 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SMART | SM00401 | 2.2E-16 | 242 | 292 | IPR000679 | Zinc finger, GATA-type |
| PROSITE profile | PS50114 | 11.662 | 242 | 278 | IPR000679 | Zinc finger, GATA-type |
| SuperFamily | SSF57716 | 7.13E-15 | 245 | 305 | No hit | No description |
| Gene3D | G3DSA:3.30.50.10 | 2.2E-14 | 246 | 279 | IPR013088 | Zinc finger, NHR/GATA-type |
| CDD | cd00202 | 1.46E-15 | 247 | 294 | No hit | No description |
| PROSITE pattern | PS00344 | 0 | 248 | 273 | IPR000679 | Zinc finger, GATA-type |
| Pfam | PF00320 | 1.4E-16 | 248 | 281 | IPR000679 | Zinc finger, GATA-type |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0008270 | Molecular Function | zinc ion binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 335 aa Download sequence Send to blast |
MAGSSLFDDN INGVLDENFD DIFRIFDFPL EDVEGNVGVE DWNVKFQGLE QPSLDFLTSY 60 SCGPSPKNGN DALKFPNGND AVKFPEVVNI STLHDETSPL KGLPVTVEAT SSRSALLHKP 120 SDVKGLRLFQ TSSPVSVLES SSSCSVENPT TVDPKTTIPV KRARNKRPRP SKSNSQFRVP 180 FFCSTSSASK ISHPSAAFES GSETSDLSET ASNPSKKKMR KKKNLSVLSG ATEMGNFSLE 240 EPVATRKCMH CEVTKTPQWR EGPMGPKTLC NACGVRYRSG RLFPEYRPAA SPTFVSSLHS 300 NSHKKVVEMR KKGSFGARMA DMDTLSSSMP TVSTG |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 165 | 169 | KRPRP |
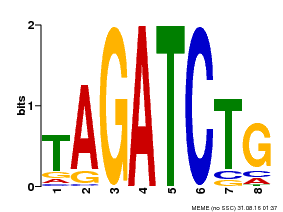
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00047 | PBM | Transfer from AT1G08000 | Download |
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | - | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_023905138.1 | 0.0 | GATA transcription factor 11 | ||||
| TrEMBL | A0A2I4EJH0 | 1e-121 | A0A2I4EJH0_JUGRE; GATA transcription factor 11-like isoform X1 | ||||
| TrEMBL | A0A2N9HVJ6 | 1e-121 | A0A2N9HVJ6_FAGSY; Uncharacterized protein | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF5758 | 28 | 49 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G08000.2 | 4e-46 | GATA transcription factor 10 | ||||



