 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | cra_locus_10206_iso_2 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Gentianales; Apocynaceae; Rauvolfioideae; Vinceae; Catharanthinae; Catharanthus
|
||||||||
| Family | ZF-HD | ||||||||
| Protein Properties | Length: 138aa MW: 15137.3 Da PI: 6.212 | ||||||||
| Description | ZF-HD family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | ZF-HD_dimer | 104.9 | 5.1e-33 | 74 | 130 | 2 | 59 |
ZF-HD_dimer 2 ekvrYkeClkNhAaslGghavDGCgEfmpsegeegtaaalkCaACgCHRnFHRrevee 59
+ v+Y+eClkNhAa++Gg+a+DGCgEfmps geegt++al+C+AC+CHRnFHR+e e
cra_locus_10206_iso_2_len_1016_ver_3 74 KMVKYRECLKNHAAAMGGNATDGCGEFMPS-GEEGTIEALTCSACNCHRNFHRKEIEG 130
679**************************9.999********************9875 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| ProDom | PD125774 | 3.0E-26 | 76 | 134 | IPR006456 | ZF-HD homeobox protein, Cys/His-rich dimerisation domain |
| Pfam | PF04770 | 6.6E-30 | 76 | 128 | IPR006456 | ZF-HD homeobox protein, Cys/His-rich dimerisation domain |
| TIGRFAMs | TIGR01566 | 3.8E-32 | 77 | 128 | IPR006456 | ZF-HD homeobox protein, Cys/His-rich dimerisation domain |
| PROSITE profile | PS51523 | 27.559 | 78 | 127 | IPR006456 | ZF-HD homeobox protein, Cys/His-rich dimerisation domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0048574 | Biological Process | long-day photoperiodism, flowering | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0042803 | Molecular Function | protein homodimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 138 aa Download sequence Send to blast |
MELPSQEGEM PIPLNSTYGG HMIHHDHSSS TVVPSQMPNS SNNNGPISSN NNNLDQDLHH 60 HNQNHHNVVV NFKKMVKYRE CLKNHAAAMG GNATDGCGEF MPSGEEGTIE ALTCSACNCH 120 RNFHRKEIEG SLPSESDX |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Putative transcription factor. Probably involved in the regulation of floral induction. {ECO:0000269|PubMed:22319055}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00258 | DAP | Transfer from AT2G02540 | Download |
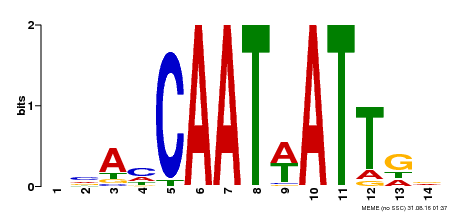
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Induced by exposure to long days. {ECO:0000269|PubMed:22319055}. | |||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_017979046.1 | 3e-52 | PREDICTED: zinc-finger homeodomain protein 4 | ||||
| Swissprot | Q9M9S0 | 8e-40 | ZHD4_ARATH; Zinc-finger homeodomain protein 4 | ||||
| TrEMBL | A0A061GFH0 | 8e-51 | A0A061GFH0_THECC; Homeobox protein 31 | ||||
| STRING | EOY25774 | 1e-51 | (Theobroma cacao) | ||||



