 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | cra_locus_102390_iso_1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Gentianales; Apocynaceae; Rauvolfioideae; Vinceae; Catharanthinae; Catharanthus
|
||||||||
| Family | WRKY | ||||||||
| Protein Properties | Length: 109aa MW: 12766.2 Da PI: 10.2773 | ||||||||
| Description | WRKY family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | WRKY | 96.5 | 1.7e-30 | 34 | 92 | 1 | 59 |
---SS-EEEEEEE--TT-SS-EEEEEE-STT---EEEEEE-SSSTTEEEEEEES--SS- CS
WRKY 1 ldDgynWrKYGqKevkgsefprsYYrCtsagCpvkkkversaedpkvveitYegeHnhe 59
ldDg++WrKYG+K+vk+s +pr+YY+C+ +gCpvkk+ver+++d ++v++tYeg Hnh+
cra_locus_102390_iso_1_len_326_ver_3 34 LDDGFKWRKYGRKMVKNSINPRNYYKCSVEGCPVKKRVERDNNDSRYVVTTYEGIHNHQ 92
59********************************************************7 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:2.20.25.80 | 2.3E-32 | 21 | 94 | IPR003657 | WRKY domain |
| SuperFamily | SSF118290 | 8.11E-28 | 27 | 94 | IPR003657 | WRKY domain |
| PROSITE profile | PS50811 | 30.171 | 29 | 94 | IPR003657 | WRKY domain |
| SMART | SM00774 | 2.1E-33 | 34 | 93 | IPR003657 | WRKY domain |
| Pfam | PF03106 | 1.4E-23 | 35 | 92 | IPR003657 | WRKY domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009867 | Biological Process | jasmonic acid mediated signaling pathway | ||||
| GO:0050832 | Biological Process | defense response to fungus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 109 aa Download sequence Send to blast |
RERSTANREN GSGREKKEIR EKVAFKTKSD VEILDDGFKW RKYGRKMVKN SINPRNYYKC 60 SVEGCPVKKR VERDNNDSRY VVTTYEGIHN HQGPMHXCQS SMIHKLPSS |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1wj2_A | 9e-25 | 24 | 95 | 7 | 78 | Probable WRKY transcription factor 4 |
| 2lex_A | 9e-25 | 24 | 95 | 7 | 78 | Probable WRKY transcription factor 4 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor. Interacts specifically with the W box (5'-(T)TGAC[CT]-3'), a frequently occurring elicitor-responsive cis-acting element (By similarity). {ECO:0000250}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00531 | DAP | Transfer from AT5G26170 | Download |
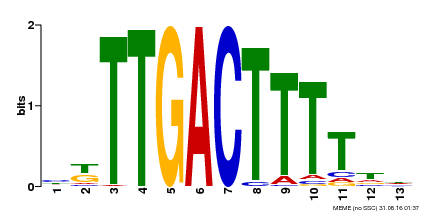
| |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_011100937.1 | 1e-50 | probable WRKY transcription factor 50 | ||||
| Swissprot | Q8VWQ5 | 2e-38 | WRK50_ARATH; Probable WRKY transcription factor 50 | ||||
| TrEMBL | A0A2G9HW23 | 5e-47 | A0A2G9HW23_9LAMI; Uncharacterized protein | ||||
| STRING | Migut.E01579.1.p | 2e-48 | (Erythranthe guttata) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Asterids | OGEA2124 | 24 | 62 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



