 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | cra_locus_108663_iso_1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Gentianales; Apocynaceae; Rauvolfioideae; Vinceae; Catharanthinae; Catharanthus
|
||||||||
| Family | TCP | ||||||||
| Protein Properties | Length: 170aa MW: 18311.3 Da PI: 10.4656 | ||||||||
| Description | TCP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | TCP | 117.6 | 1.6e-36 | 76 | 147 | 2 | 73 |
TCP 2 agkkdrhskihTkvggRdRRvRlsaecaarfFdLqdeLGfdkdsktieWLlqqakpaikeltgtssssasec 73
+ ++++++++hTkv+gR+RR+R++a caar+F+L++eLG+++d++ti WLl+qa+p+i+++tgt++ +a ++
cra_locus_108663_iso_1_len_509_ver_3 76 PLRRSSTKDRHTKVEGRGRRIRMPAACAARVFQLTKELGHKSDGETIRWLLEQAEPSIIAATGTGTVPAIAM 147
57899***********************************************************88887443 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF03634 | 3.9E-30 | 81 | 153 | IPR005333 | Transcription factor, TCP |
| PROSITE profile | PS51369 | 26.49 | 82 | 136 | IPR017887 | Transcription factor TCP subgroup |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0008361 | Biological Process | regulation of cell size | ||||
| GO:0048364 | Biological Process | root development | ||||
| GO:1900056 | Biological Process | negative regulation of leaf senescence | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 170 aa Download sequence Send to blast |
INPETLRLME SRNFYQSSGL PPQLTMAQPT SIPIISLKEE PELEEIKPSL LMGSTLPMPM 60 PLQMRQPAMA PPPPPPLRRS STKDRHTKVE GRGRRIRMPA ACAARVFQLT KELGHKSDGE 120 TIRWLLEQAE PSIIAATGTG TVPAIAMSVN GTLKIPTTSP TSAPSAAAAA |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00667 | PBM | Transfer from GSVIVG01027588001 | Download |
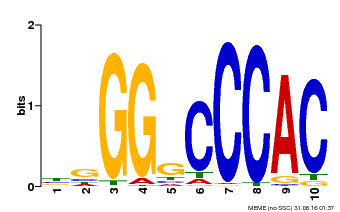
| |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_027103361.1 | 3e-61 | transcription factor TCP9-like | ||||
| Swissprot | O64647 | 1e-51 | TCP9_ARATH; Transcription factor TCP9 | ||||
| TrEMBL | A0A068UKS1 | 1e-60 | A0A068UKS1_COFCA; Uncharacterized protein | ||||
| STRING | VIT_15s0048g01150.t01 | 1e-56 | (Vitis vinifera) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Asterids | OGEA248 | 24 | 201 |



