 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | cra_locus_11227_iso_4 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Gentianales; Apocynaceae; Rauvolfioideae; Vinceae; Catharanthinae; Catharanthus
|
||||||||
| Family | HD-ZIP | ||||||||
| Protein Properties | Length: 244aa MW: 28403.8 Da PI: 7.6177 | ||||||||
| Description | HD-ZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Homeobox | 57.1 | 3e-18 | 24 | 78 | 2 | 56 |
T--SS--HHHHHHHHHHHHHSSS--HHHHHHHHHHCTS-HHHHHHHHHHHHHHHH CS
Homeobox 2 rkRttftkeqleeLeelFeknrypsaeereeLAkklgLterqVkvWFqNrRakek 56
rk+ +++ eq+++Le+ Fe +++ e++ +LA+ lgL+ rq+ +WFqNrRa++k
cra_locus_11227_iso_4_len_1532_ver_3 24 RKKGRLNMEQVKTLEKNFELGNKLEPERKMQLARALGLQPRQIAIWFQNRRARWK 78
67778999**********************************************9 PP
| |||||||
| 2 | HD-ZIP_I/II | 129.5 | 1.3e-41 | 24 | 116 | 1 | 93 |
HD-ZIP_I/II 1 ekkrrlskeqvklLEesFeeeekLeperKvelareLglqprqvavWFqnrRARtktkqlEkdyeaLkraydal 73
+kk rl+ eqvk+LE++Fe +kLeperK++lar+Lglqprq+a+WFqnrRAR+ktkqlEkdy+ Lkr+++a+
cra_locus_11227_iso_4_len_1532_ver_3 24 RKKGRLNMEQVKTLEKNFELGNKLEPERKMQLARALGLQPRQIAIWFQNRRARWKTKQLEKDYDILKRQFEAI 96
6999********************************************************************* PP
HD-ZIP_I/II 74 keenerLekeveeLreelke 93
k+en++L++++++L++e+++
cra_locus_11227_iso_4_len_1532_ver_3 97 KAENDALQAQNQKLHAEIMA 116
***************99975 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF46689 | 6.42E-19 | 15 | 82 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS50071 | 17.346 | 20 | 80 | IPR001356 | Homeobox domain |
| SMART | SM00389 | 1.6E-17 | 22 | 84 | IPR001356 | Homeobox domain |
| Pfam | PF00046 | 1.3E-15 | 24 | 78 | IPR001356 | Homeobox domain |
| CDD | cd00086 | 1.34E-14 | 24 | 81 | No hit | No description |
| Gene3D | G3DSA:1.10.10.60 | 1.0E-19 | 25 | 87 | IPR009057 | Homeodomain-like |
| PRINTS | PR00031 | 8.7E-6 | 51 | 60 | IPR000047 | Helix-turn-helix motif |
| PROSITE pattern | PS00027 | 0 | 55 | 78 | IPR017970 | Homeobox, conserved site |
| PRINTS | PR00031 | 8.7E-6 | 60 | 76 | IPR000047 | Helix-turn-helix motif |
| Pfam | PF02183 | 5.1E-17 | 80 | 120 | IPR003106 | Leucine zipper, homeobox-associated |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009744 | Biological Process | response to sucrose | ||||
| GO:0048826 | Biological Process | cotyledon morphogenesis | ||||
| GO:0080022 | Biological Process | primary root development | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 244 aa Download sequence Send to blast |
MESEAAVATE ERMIYQMMDH NWERKKGRLN MEQVKTLEKN FELGNKLEPE RKMQLARALG 60 LQPRQIAIWF QNRRARWKTK QLEKDYDILK RQFEAIKAEN DALQAQNQKL HAEIMALKNR 120 EPTESINLNK ETEGSCSNRS ENSSDNIKLD ISRTTPAIDS HPQTSRPFFP SSLIRPNTAQ 180 QLFQTTSSSS RPPPADHHHL IQCQNNNNNN PTATVKEESS LCNMFCTIDD QTGFWPWLEQ 240 HHFN |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 72 | 80 | RRARWKTKQ |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor that may act in the sucrose-signaling pathway. {ECO:0000269|PubMed:11292072}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00225 | DAP | Transfer from AT1G69780 | Download |
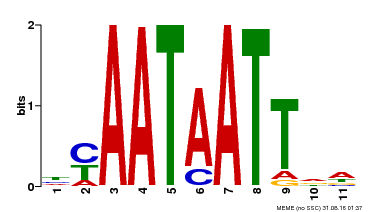
| |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_002520111.1 | 1e-115 | homeobox-leucine zipper protein ATHB-13 | ||||
| Refseq | XP_015575250.1 | 1e-115 | homeobox-leucine zipper protein ATHB-13 | ||||
| Swissprot | Q8LC03 | 7e-93 | ATB13_ARATH; Homeobox-leucine zipper protein ATHB-13 | ||||
| TrEMBL | A1DR78 | 1e-161 | A1DR78_CATRO; DNA-binding protein (Fragment) | ||||
| STRING | XP_002520111.1 | 1e-114 | (Ricinus communis) | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



