 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | cra_locus_12315_iso_3 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Gentianales; Apocynaceae; Rauvolfioideae; Vinceae; Catharanthinae; Catharanthus
|
||||||||
| Family | bHLH | ||||||||
| Protein Properties | Length: 507aa MW: 55715.1 Da PI: 7.1546 | ||||||||
| Description | bHLH family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | HLH | 49.8 | 6.1e-16 | 326 | 372 | 4 | 55 |
HHHHHHHHHHHHHHHHHHHHHCTSCCC...TTS-STCHHHHHHHHHHHHHHH CS
HLH 4 ahnerErrRRdriNsafeeLrellPkaskapskKlsKaeiLekAveYIksLq 55
hn ErrRRdriN+++ L+ellP++ K +Ka++L +A+eY+ksLq
cra_locus_12315_iso_3_len_2534_ver_3 326 VHNLSERRRRDRINEKMRALQELLPHS-----NKSDKASMLDEAIEYMKSLQ 372
6*************************7.....59*****************9 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF47459 | 1.09E-19 | 320 | 384 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| PROSITE profile | PS50888 | 17.815 | 322 | 371 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| CDD | cd00083 | 4.64E-17 | 325 | 376 | No hit | No description |
| Pfam | PF00010 | 2.0E-13 | 326 | 372 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene3D | G3DSA:4.10.280.10 | 3.6E-19 | 326 | 381 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SMART | SM00353 | 8.3E-17 | 328 | 377 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009585 | Biological Process | red, far-red light phototransduction | ||||
| GO:0009693 | Biological Process | ethylene biosynthetic process | ||||
| GO:0009704 | Biological Process | de-etiolation | ||||
| GO:0010161 | Biological Process | red light signaling pathway | ||||
| GO:0010244 | Biological Process | response to low fluence blue light stimulus by blue low-fluence system | ||||
| GO:0010600 | Biological Process | regulation of auxin biosynthetic process | ||||
| GO:0010928 | Biological Process | regulation of auxin mediated signaling pathway | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 507 aa Download sequence Send to blast |
MYPCFPDWNI EAELPVLHQK KPFGLEHELV ELLWENGQVV LHSQTHRKSG PAGVPDHPFD 60 SSTRQLNNNN NSNNKQATTS CGNPTATLIQ DNDTVSWIHC QVDDPFDKEF SSNFLPVIPL 120 PTPQLEDPPH KQQIQHCRTH PSTNFGPNPL PAPRFQSDSN NVRGGVSKCP NVLLKADLGS 180 SNSGPRPSSH MIIGEVREYS MRTVGSSHCG SNQVAMDADT SRASSCGIGN NDLSAAAVVK 240 DYEGKLGSQN YERGERETHE LANTSSSGGS GTSFGRTCTA TDSLKRKSRD AEDSECQSEA 300 AVLESEARKK PASKSGIARK SRAAEVHNLS ERRRRDRINE KMRALQELLP HSNKSDKASM 360 LDEAIEYMKS LQLQLQMMWM RSGMAPLMFP GVQHYMSRLG MGIGPPTLPP VPNPMHLSRL 420 PLVDQTMTVA SAPNQAALCP TSVINPINYQ NQLQNSNFSE QYATYMGFHH PMQTSSQPIN 480 VFGFNSNMTQ QNHNFAPPSN TNGSSAG |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 330 | 335 | ERRRRD |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00606 | ChIP-seq | Transfer from AT2G43010 | Download |
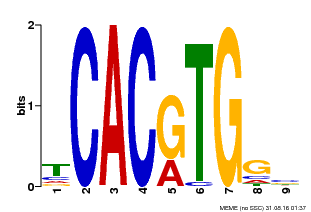
| |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_027061311.1 | 1e-171 | transcription factor PHYTOCHROME INTERACTING FACTOR-LIKE 13-like isoform X1 | ||||
| Refseq | XP_027061312.1 | 1e-171 | transcription factor PHYTOCHROME INTERACTING FACTOR-LIKE 13-like isoform X1 | ||||
| Refseq | XP_027061313.1 | 1e-171 | transcription factor PHYTOCHROME INTERACTING FACTOR-LIKE 13-like isoform X1 | ||||
| TrEMBL | A0A0N7HUP6 | 0.0 | A0A0N7HUP6_CATRO; Phytochrome interacting factor 5-like protein | ||||
| STRING | XP_006485007.1 | 1e-166 | (Citrus sinensis) | ||||
| STRING | XP_009757612.1 | 1e-166 | (Nicotiana sylvestris) | ||||



