 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | cra_locus_12712_iso_2 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Gentianales; Apocynaceae; Rauvolfioideae; Vinceae; Catharanthinae; Catharanthus
|
||||||||
| Family | NAC | ||||||||
| Protein Properties | Length: 100aa MW: 11842.8 Da PI: 7.817 | ||||||||
| Description | NAC family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | NAM | 61.7 | 2.3e-19 | 26 | 74 | 3 | 52 |
NAM 3 pGfrFhPtdeelvveyLkkkvegkkleleevikevdiykvePwdLpkkvk 52
pGfrFhPtdeelv +yL++kve++++++ e ik++diyk++Pw+Lp+k +
cra_locus_12712_iso_2_len_297_ver_3 26 PGFRFHPTDEELVGFYLRRKVEKRPISI-ELIKQIDIYKHDPWNLPSKSY 74
9***************************.89***************5433 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51005 | 19.506 | 24 | 100 | IPR003441 | NAC domain |
| SuperFamily | SSF101941 | 3.4E-18 | 25 | 74 | IPR003441 | NAC domain |
| Pfam | PF02365 | 2.4E-9 | 26 | 97 | IPR003441 | NAC domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0005992 | Biological Process | trehalose biosynthetic process | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0006561 | Biological Process | proline biosynthetic process | ||||
| GO:0009718 | Biological Process | anthocyanin-containing compound biosynthetic process | ||||
| GO:0010120 | Biological Process | camalexin biosynthetic process | ||||
| GO:0042538 | Biological Process | hyperosmotic salinity response | ||||
| GO:1900056 | Biological Process | negative regulation of leaf senescence | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 100 aa Download sequence Send to blast |
XRVEDMNGGG VIVSGGKDEE DDVPLPGFRF HPTDEELVGF YLRRKVEKRP ISIELIKQID 60 IYKHDPWNLP SKSYAHHVLI SWLDRPRPLL XYPKKYFYYX |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor that binds to the 5'- RRYGCCGT-3' consensus core sequence. Central longevity regulator. Negative regulator of leaf senescence. Modulates cellular H(2)O(2) levels and enhances tolerance to various abiotic stresses through the regulation of DREB2A. {ECO:0000269|PubMed:22345491}. | |||||
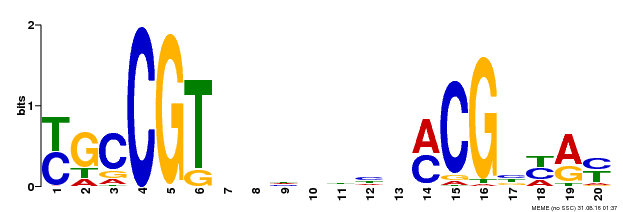
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00310 | DAP | Transfer from AT2G43000 | Download |
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Up-regulated by H(2)O(2), paraquat, ozone, 3-aminotriazole and salt stress. {ECO:0000269|PubMed:22345491}. | |||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_027063813.1 | 1e-31 | transcription factor JUNGBRUNNEN 1-like | ||||
| Swissprot | Q9SK55 | 2e-23 | NAC42_ARATH; Transcription factor JUNGBRUNNEN 1 | ||||
| TrEMBL | A0A068UZ64 | 2e-28 | A0A068UZ64_COFCA; Uncharacterized protein | ||||
| STRING | LPERR12G16500.1 | 5e-27 | (Leersia perrieri) | ||||



