 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | cra_locus_1311_iso_7 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Gentianales; Apocynaceae; Rauvolfioideae; Vinceae; Catharanthinae; Catharanthus
|
||||||||
| Family | WRKY | ||||||||
| Protein Properties | Length: 343aa MW: 37986.2 Da PI: 7.8177 | ||||||||
| Description | WRKY family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | WRKY | 89.7 | 2.3e-28 | 182 | 241 | 1 | 59 |
---SS-EEEEEEE--TT-SS-EEEEEE-ST.T---EEEEEE-SSSTTEEEEEEES--SS- CS
WRKY 1 ldDgynWrKYGqKevkgsefprsYYrCtsa.gCpvkkkversaedpkvveitYegeHnhe 59
++Dgy+WrKYGqK+ ++++ pr+Y++C++a +Cpvkkkv+ + ed+++++ tYegeHnh+
cra_locus_1311_iso_7_len_1818_ver_3 182 VKDGYQWRKYGQKVTRDNPSPRAYFKCSFApSCPVKKKVKEAXEDQSILVATYEGEHNHP 241
58***************************99****************************8 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:2.20.25.80 | 2.3E-28 | 172 | 243 | IPR003657 | WRKY domain |
| PROSITE profile | PS50811 | 25.901 | 177 | 243 | IPR003657 | WRKY domain |
| SuperFamily | SSF118290 | 4.05E-24 | 179 | 243 | IPR003657 | WRKY domain |
| SMART | SM00774 | 2.1E-30 | 182 | 242 | IPR003657 | WRKY domain |
| Pfam | PF03106 | 4.6E-23 | 183 | 241 | IPR003657 | WRKY domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0002237 | Biological Process | response to molecule of bacterial origin | ||||
| GO:0009611 | Biological Process | response to wounding | ||||
| GO:0009751 | Biological Process | response to salicylic acid | ||||
| GO:0010200 | Biological Process | response to chitin | ||||
| GO:0042742 | Biological Process | defense response to bacterium | ||||
| GO:0045892 | Biological Process | negative regulation of transcription, DNA-templated | ||||
| GO:0050691 | Biological Process | regulation of defense response to virus by host | ||||
| GO:0050832 | Biological Process | defense response to fungus | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| GO:0044212 | Molecular Function | transcription regulatory region DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 343 aa Download sequence Send to blast |
MEYTSLIDTS LDLNVKPLRL IDNGSLKQEV RSNFIDQLGR KMPLVKEEEN HGALMEELNR 60 VSAENQKLTQ MLTVMCENYD ALRKHLMDYM DKNPSSGGGG TDDINNNNNN NNASKKRKLA 120 ESSSNINNDT IVDAAVATTT NGPSESSSSD EGSCKKPREE HIKSKISRVY VRTEASDTSL 180 IVKDGYQWRK YGQKVTRDNP SPRAYFKCSF APSCPVKKKV KEAXEDQSIL VATYEGEHNH 240 PHPSKMENTS TTSKCCVTTT IGSVPCSTSL SSSGGGPTIT LDLTKQKSHE TNPNPRKLES 300 PEFQHFLIEQ MASSLTKDPS FKAALAAAIS GKFLQHNNQT EKW |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 2ayd_A | 8e-19 | 167 | 243 | 1 | 74 | WRKY transcription factor 1 |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00251 | DAP | Transfer from AT1G80840 | Download |
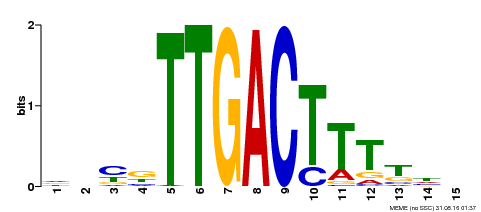
| |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_027167435.1 | 1e-159 | probable WRKY transcription factor 40 | ||||
| TrEMBL | A0A2U9IY44 | 0.0 | A0A2U9IY44_CATRO; WRKY DNA-binding protein 40 | ||||
| STRING | cassava4.1_011696m | 1e-127 | (Manihot esculenta) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Asterids | OGEA2855 | 24 | 50 |



