 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | cra_locus_13167_iso_2 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Gentianales; Apocynaceae; Rauvolfioideae; Vinceae; Catharanthinae; Catharanthus
|
||||||||
| Family | CAMTA | ||||||||
| Protein Properties | Length: 1061aa MW: 117811 Da PI: 5.8833 | ||||||||
| Description | CAMTA family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | CG-1 | 117.2 | 8.3e-37 | 2 | 73 | 47 | 118 |
CG-1 47 vryfrkDGyswkkkkdgktvrEdhekLKvggvevlycyYahseenptfqrrcywlLeeelekivlvhylevk 118
+ryfrkDG++w+kk+dgktv+E+he+LK g+v+vl+cyYah+een++fqrr+yw+Leee+++ivlvhy+evk
cra_locus_13167_iso_2_len_3478_ver_3 2 LRYFRKDGHNWRKKRDGKTVKEAHERLKAGSVDVLHCYYAHGEENENFQRRSYWMLEEERSHIVLVHYREVK 73
79*******************************************************************986 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SMART | SM01076 | 7.0E-29 | 1 | 73 | IPR005559 | CG-1 DNA-binding domain |
| PROSITE profile | PS51437 | 55.349 | 1 | 78 | IPR005559 | CG-1 DNA-binding domain |
| Pfam | PF03859 | 4.5E-30 | 2 | 71 | IPR005559 | CG-1 DNA-binding domain |
| SuperFamily | SSF81296 | 3.36E-11 | 470 | 556 | IPR014756 | Immunoglobulin E-set |
| SuperFamily | SSF48403 | 1.71E-16 | 651 | 767 | IPR020683 | Ankyrin repeat-containing domain |
| CDD | cd00204 | 5.55E-13 | 653 | 765 | No hit | No description |
| Gene3D | G3DSA:1.25.40.20 | 1.4E-16 | 653 | 768 | IPR020683 | Ankyrin repeat-containing domain |
| PROSITE profile | PS50297 | 16.626 | 673 | 777 | IPR020683 | Ankyrin repeat-containing domain |
| Pfam | PF12796 | 1.5E-7 | 678 | 768 | IPR020683 | Ankyrin repeat-containing domain |
| PROSITE profile | PS50088 | 9.458 | 706 | 738 | IPR002110 | Ankyrin repeat |
| SMART | SM00248 | 0.0018 | 706 | 735 | IPR002110 | Ankyrin repeat |
| SMART | SM00248 | 250 | 745 | 774 | IPR002110 | Ankyrin repeat |
| SuperFamily | SSF52540 | 1.73E-8 | 877 | 932 | IPR027417 | P-loop containing nucleoside triphosphate hydrolase |
| SMART | SM00015 | 0.11 | 881 | 903 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 8.261 | 883 | 911 | IPR000048 | IQ motif, EF-hand binding site |
| Pfam | PF00612 | 0.002 | 883 | 902 | IPR000048 | IQ motif, EF-hand binding site |
| SMART | SM00015 | 7.5E-4 | 904 | 926 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 9.578 | 905 | 929 | IPR000048 | IQ motif, EF-hand binding site |
| Pfam | PF00612 | 8.1E-5 | 907 | 926 | IPR000048 | IQ motif, EF-hand binding site |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009409 | Biological Process | response to cold | ||||
| GO:0010150 | Biological Process | leaf senescence | ||||
| GO:0042742 | Biological Process | defense response to bacterium | ||||
| GO:0050832 | Biological Process | defense response to fungus | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0005516 | Molecular Function | calmodulin binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 1061 aa Download sequence Send to blast |
XLRYFRKDGH NWRKKRDGKT VKEAHERLKA GSVDVLHCYY AHGEENENFQ RRSYWMLEEE 60 RSHIVLVHYR EVKGNRTNLI RSREATEPIP DAQETEDDIR NSEAGSCSTS KFHPYDYQVA 120 SQVTDTASLN SAQASEYGDA ESAYNHQSSS SFHTYRDLQP PVMQKAEDRH SLPVPSIELV 180 LLVLLFFSFV CVAIVVAFGD EKSKFLLTDT YQVQFPGPPN MGFDPNATST GAPYMPEGQH 240 DFSTWRNVIE SGGLAGQPIN FQTSQVTNSS MMHGLLQQVL TDSLGTEPVS GSRTDGLGQW 300 QTSEGDPLYI SKWPMDQKLH PDSANKLVAS SSEDNIAGLH NSLAPYNVLS ANQNTPQNDL 360 PVHLGGANAG NSSKPAWNSN QTIEGKADYP TMKQVLLDGV LEGGLKKLDS FDRWMSRELG 420 DVNDSHIQSS STNYWESVGN EDDNSNITPP EQIDSYMMGP SLSQDQLFSI IDFAPNWAFS 480 GSEVKVXXXX XXXRAXEEAE KCNWACMFGE LEVPAEVIAN GVLRCSTPLH EPGRVPFYVT 540 CSNRCACSEV REFEFRISSV QNEDVAVMSS VSSEESHLLI RFTKLLTAGS LSNPTNVPGA 600 AGGLSHLTNK VDSSVLDDEN EWACIQNLIS EDKFSVKKVK DQLLQKLLRD KLHLWLIQKV 660 AEDGKGPLIL DEGGQGVLHF AAALGYDWAI PPTIAGGVSV NFRDVNGWTA LHWAASYGRE 720 RTVASLISLD ANPGALTDPT PKYPSGRTPA DLAYSNGHKG IAGYLAESAL SSHLSTIELK 780 DKKDGQDVEN SELKAVQTIT ERTATPSDYG DLPHGLSMKD SLAALRNATQ AAARIHQVFR 840 VQSFQRKQIK EYGDGGSGIS DERALSLLAM RSNRAGHPAE PVHAAAIRIQ NKFRGWKGRK 900 EFLLIRQRII KIQAHVRGHQ VRKNYKKIIW SVGILDKVIL RWRRKGSGLR GFKSEAITEG 960 SGGVERSSKE DDYDFLKEGR KQTEDRLQKA LARVKSMVQY PEARDQYRRL LNVVSEMQET 1020 KTKYDRLLNN SDETAADFDD DLIDIDALLE DDTSMAASSS T |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription activator that binds to the DNA consensus sequence 5'-[ACG]CGCG[GTC]-3'. Binds calmodulin in a calcium-dependent manner in vitro (PubMed:12218065). Regulates transcriptional activity in response to calcium signals (Probable). Involved in freezing tolerance in association with CAMTA1 and CAMTA2 (PubMed:23581962). Required for the cold-induced expression of DREB1B/CBF1, DREB1C/CBF2, ZAT12 and GOLS3 (PubMed:19270186). Involved in response to cold. Contributes together with CAMTA5 to the positive regulation of the cold-induced expression of DREB1A/CBF3, DREB1B/CBF1 and DREB1C/CBF2 (PubMed:28351986). Involved together with CAMTA2 and CAMTA4 in the positive regulation of a general stress response (GSR) (PubMed:25039701). Involved in the regulation of GSR amplitude downstream of MEKK1 (PubMed:25157030). Involved in the regulation of a set of genes involved in defense responses against pathogens (PubMed:18298954). Involved in the regulation of both basal resistance and systemic acquired resistance (SAR) (PubMed:21900483). Acts as negative regulator of plant immunity (PubMed:19122675, PubMed:21900483, PubMed:22345509, PubMed:28407487). Binds to the promoter of the defense-related gene EDS1 and represses its expression (PubMed:19122675). Binds to the promoter of the defense-related gene NDR1 and represses its expression (PubMed:22345509). Involved in defense against insects (PubMed:23072934, PubMed:22371088). Required for tolerance to the generalist herbivore Trichoplusia ni, and contributes to the positive regulation of genes associated with glucosinolate metabolism (PubMed:23072934). Required for tolerance to Bradysia impatiens larvae. Mediates herbivore-induced wound response (PubMed:22371088). Required for wound-induced jasmonate accumulation (PubMed:23072934, PubMed:22371088). Involved in the regulation of ethylene-induced senescence by binding to the promoter of the senescence-inducer gene EIN3 and repressing its expression (PubMed:22345509). {ECO:0000269|PubMed:12218065, ECO:0000269|PubMed:18298954, ECO:0000269|PubMed:19122675, ECO:0000269|PubMed:19270186, ECO:0000269|PubMed:21900483, ECO:0000269|PubMed:22345509, ECO:0000269|PubMed:22371088, ECO:0000269|PubMed:23072934, ECO:0000269|PubMed:23581962, ECO:0000269|PubMed:25039701, ECO:0000269|PubMed:25157030, ECO:0000269|PubMed:28351986, ECO:0000269|PubMed:28407487, ECO:0000305|PubMed:11925432}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00042 | PBM | Transfer from AT2G22300 | Download |
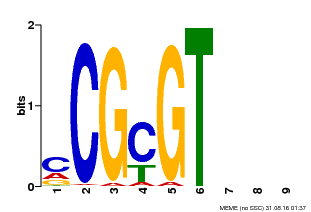
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: By heat shock, UVB, salt, wounding, ethylene and methyl jasmonate (PubMed:11162426, PubMed:12218065). Induced by infection with the fungal pathogen Golovinomyces cichoracearum (powdery mildew) and the bacterial pathogen Pseudomonas syringae pv tomato strain DC3000 (PubMed:22345509). {ECO:0000269|PubMed:11162426, ECO:0000269|PubMed:12218065, ECO:0000269|PubMed:22345509}. | |||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_027078238.1 | 0.0 | calmodulin-binding transcription activator 3-like | ||||
| Swissprot | Q8GSA7 | 0.0 | CMTA3_ARATH; Calmodulin-binding transcription activator 3 | ||||
| TrEMBL | A0A068TZG3 | 0.0 | A0A068TZG3_COFCA; Uncharacterized protein | ||||
| STRING | XP_009613615.1 | 0.0 | (Nicotiana tomentosiformis) | ||||



