 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | cra_locus_14079_iso_1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Gentianales; Apocynaceae; Rauvolfioideae; Vinceae; Catharanthinae; Catharanthus
|
||||||||
| Family | MYB_related | ||||||||
| Protein Properties | Length: 76aa MW: 8753.89 Da PI: 9.387 | ||||||||
| Description | MYB_related family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 56.8 | 5.3e-18 | 14 | 61 | 1 | 48 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqkyl 48
+g+WT+eEd++l+ +++ +G g+W++ ++ g+ R++k+c++rw +yl
cra_locus_14079_iso_1_len_262_ver_3 14 KGAWTKEEDDRLIAYIRAHGEGCWRSLPKAAGLLRCGKSCRLRWINYL 61
79******************************99************97 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:1.10.10.60 | 1.8E-24 | 5 | 69 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS51294 | 22.913 | 9 | 65 | IPR017930 | Myb domain |
| SMART | SM00717 | 1.9E-13 | 13 | 63 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 2.9E-16 | 14 | 61 | IPR001005 | SANT/Myb domain |
| SuperFamily | SSF46689 | 3.19E-17 | 15 | 71 | IPR009057 | Homeodomain-like |
| CDD | cd00167 | 2.59E-10 | 16 | 61 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009751 | Biological Process | response to salicylic acid | ||||
| GO:0009753 | Biological Process | response to jasmonic acid | ||||
| GO:0010224 | Biological Process | response to UV-B | ||||
| GO:0045892 | Biological Process | negative regulation of transcription, DNA-templated | ||||
| GO:0090379 | Biological Process | secondary cell wall biogenesis involved in seed trichome differentiation | ||||
| GO:1903086 | Biological Process | negative regulation of sinapate ester biosynthetic process | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 76 aa Download sequence Send to blast |
MGRSPCCEKA HTNKGAWTKE EDDRLIAYIR AHGEGCWRSL PKAAGLLRCG KSCRLRWINY 60 LRPDLKRGSL XLLEDY |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor. {ECO:0000305}. | |||||
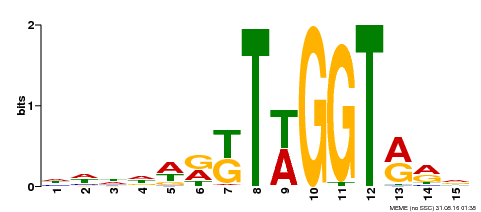
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00479 | DAP | Transfer from AT4G38620 | Download |
| |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_022952474.1 | 2e-45 | myb-related protein 308-like | ||||
| Swissprot | P81393 | 7e-46 | MYB08_ANTMA; Myb-related protein 308 | ||||
| TrEMBL | A0A2U1LUN3 | 2e-44 | A0A2U1LUN3_ARTAN; Myb-related protein Hv1 | ||||
| STRING | Gorai.008G035700.1 | 4e-44 | (Gossypium raimondii) | ||||
| STRING | EMJ01802 | 4e-44 | (Prunus persica) | ||||
| STRING | XP_004289866.1 | 5e-44 | (Fragaria vesca) | ||||
| STRING | XP_010090332.1 | 5e-44 | (Morus notabilis) | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



