 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | cra_locus_14743_iso_2 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Gentianales; Apocynaceae; Rauvolfioideae; Vinceae; Catharanthinae; Catharanthus
|
||||||||
| Family | Trihelix | ||||||||
| Protein Properties | Length: 429aa MW: 49771.2 Da PI: 5.3594 | ||||||||
| Description | Trihelix family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | trihelix | 94.7 | 8.7e-30 | 277 | 362 | 1 | 86 |
trihelix 1 rWtkqevlaLiearremeerlrrgklkkplWeevskkmrergferspkqCkekwenlnkrykkikegekkrts 73
rW+k ev aLi++r++ e+++++ lk+ lWee+s+ m++ gf+rs+k+Ckekwen+nk++kk ke+ +k++
cra_locus_14743_iso_2_len_1453_ver_3 277 RWPKAEVEALIQIRSRFETNFQEPGLKGLLWEEISNEMAKMGFQRSAKRCKEKWENINKYFKKSKENPIKERR 349
8************************************************************************ PP
trihelix 74 essstcpyfdqle 86
++ +tc yfdql+
cra_locus_14743_iso_2_len_1453_ver_3 350 QKLKTCEYFDQLD 362
************8 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:1.10.10.60 | 1.2E-5 | 267 | 333 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS50090 | 7.48 | 270 | 334 | IPR017877 | Myb-like domain |
| SuperFamily | SSF46689 | 1.79E-5 | 272 | 341 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 0.0015 | 274 | 336 | IPR001005 | SANT/Myb domain |
| CDD | cd12203 | 1.41E-24 | 276 | 341 | No hit | No description |
| Pfam | PF13837 | 1.5E-21 | 276 | 364 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 429 aa Download sequence Send to blast |
MKCCRKQFCP DISEAKNIER GIIEAKDKDQ EEQPEMNQNQ NHHEDYNHDF DEETVQNSCL 60 EAGKSRLFGE LEAICGHASS SSCIGKSNHH HKSAAGSLII NGGETLAEGT CLAPPALDQS 120 NHLNNASSDQ TLDEVEEDEP FSRMVKKKRK KREKMKDDEI SSMAQLFERL VKQLMDHQEI 180 LYKKFVDMIE RIDKERKDRE EAWRKQESEV ASKAHERTLL ASSSTKESSS NVFEYLERIT 240 GKSMNLISPP IRDHEPPLEL SNNRSRNNVD QNVMSKRWPK AEVEALIQIR SRFETNFQEP 300 GLKGLLWEEI SNEMAKMGFQ RSAKRCKEKW ENINKYFKKS KENPIKERRQ KLKTCEYFDQ 360 LDQLYSKAQE KGYYNIKEVD FEGSSNYGKF QVKKNHDDNN HEEADHEDLV GQTEEGEEED 420 NKGESGHEE |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 145 | 151 | KKKRKKR |
| 2 | 146 | 150 | KKRKK |
| 3 | 148 | 153 | RKKREK |
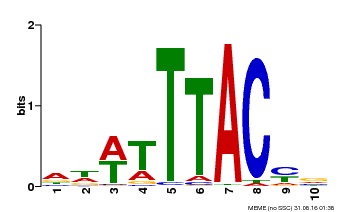
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00651 | PBM | Transfer from LOC_Os02g01380 | Download |
| |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_027084142.1 | 2e-85 | trihelix transcription factor GT-2-like | ||||



