 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | cra_locus_16284_iso_3 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Gentianales; Apocynaceae; Rauvolfioideae; Vinceae; Catharanthinae; Catharanthus
|
||||||||
| Family | WRKY | ||||||||
| Protein Properties | Length: 328aa MW: 36932.4 Da PI: 6.5109 | ||||||||
| Description | WRKY family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | WRKY | 76.1 | 4.1e-24 | 117 | 177 | 1 | 59 |
---SS-EEEEEEE--TT-SS-EEEEEE-ST..T---EEEEEE-SSSTTEEEEEEES--SS- CS
WRKY 1 ldDgynWrKYGqKevkgsefprsYYrCtsa..gCpvkkkversaedpkvveitYegeHnhe 59
++D+y+WrKYGqK++ s+fpr Y+rCt++ gC+++k+v+ +++p + ++tY g+H+++
cra_locus_16284_iso_3_len_1580_ver_3 117 MEDEYAWRKYGQKDILRSNFPRCYFRCTHKneGCKATKQVQIVTKNPLMYQTTYFGQHTCN 177
58***************************9999**************************96 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:2.20.25.80 | 4.9E-23 | 104 | 177 | IPR003657 | WRKY domain |
| PROSITE profile | PS50811 | 17.89 | 112 | 174 | IPR003657 | WRKY domain |
| SuperFamily | SSF118290 | 1.44E-22 | 115 | 177 | IPR003657 | WRKY domain |
| SMART | SM00774 | 2.3E-32 | 117 | 178 | IPR003657 | WRKY domain |
| Pfam | PF03106 | 1.6E-23 | 118 | 176 | IPR003657 | WRKY domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009862 | Biological Process | systemic acquired resistance, salicylic acid mediated signaling pathway | ||||
| GO:0009864 | Biological Process | induced systemic resistance, jasmonic acid mediated signaling pathway | ||||
| GO:0010200 | Biological Process | response to chitin | ||||
| GO:0045892 | Biological Process | negative regulation of transcription, DNA-templated | ||||
| GO:0050832 | Biological Process | defense response to fungus | ||||
| GO:1900056 | Biological Process | negative regulation of leaf senescence | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 328 aa Download sequence Send to blast |
MAWHDRKKLL EEILLKGKES ATKLQTLLHQ KPNNSPYDVV SAAELSVQIF RSFTETLAVL 60 GPDDIRQIVA ADGMARSSSS EITDGFIRKK SAGVKYRKGS YKRRNVSETE TKYSSTMEDE 120 YAWRKYGQKD ILRSNFPRCY FRCTHKNEGC KATKQVQIVT KNPLMYQTTY FGQHTCNDHL 180 LMRAPHHDII QEISSDPMDS CLLSFQTTVN NIPSSSSNNQ AIKPLPKQVI VAVTKEDSDD 240 FSSDGKSLPS HSTLLQELVT SGSSSRSPAN LIPSKTRSAY LEEVISGNYS CSSNNTLDDL 300 DMDIEVFNTK FGDLDNNFCF DEEISDLF |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 6ir8_A | 4e-15 | 117 | 176 | 8 | 68 | OsWRKY45 |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00408 | DAP | Transfer from AT3G56400 | Download |
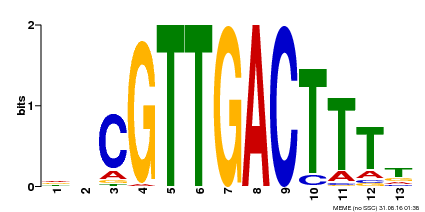
| |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_027104131.1 | 4e-76 | WRKY DNA-binding transcription factor 70-like | ||||
| TrEMBL | F6KW33 | 0.0 | F6KW33_CATRO; WRKY1 | ||||



