 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | cra_locus_18518_iso_1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Gentianales; Apocynaceae; Rauvolfioideae; Vinceae; Catharanthinae; Catharanthus
|
||||||||
| Family | MYB | ||||||||
| Protein Properties | Length: 354aa MW: 39696.4 Da PI: 6.5091 | ||||||||
| Description | MYB family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 54.2 | 3.4e-17 | 14 | 62 | 1 | 48 |
TSSS-HHHHHHHHHHHHHTTTT.-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGgg.tWktIartmgkgRtlkqcksrwqkyl 48
+g+W++eEd +l+ +++q+G+g +W + ++++g++R++k+c++rw++yl
cra_locus_18518_iso_1_len_1237_ver_3 14 KGPWSPEEDAKLKAYIEQHGTGgNWIALPQKIGLKRCGKSCRLRWLNYL 62
79*********************************************97 PP
| |||||||
| 2 | Myb_DNA-binding | 37.5 | 5.5e-12 | 69 | 111 | 2 | 46 |
SSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHH CS
Myb_DNA-binding 2 grWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqk 46
g +++eEd ++ + G++ W+ Ia+ ++ gRt++++k++w++
cra_locus_18518_iso_1_len_1237_ver_3 69 GGFSEEEDNIICSLYISIGSR-WSIIAAQLP-GRTDNDIKNYWNT 111
569******************.*********.***********97 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51294 | 14.783 | 9 | 62 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 1.06E-26 | 11 | 109 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 3.2E-13 | 13 | 64 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 9.8E-16 | 14 | 62 | IPR001005 | SANT/Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 5.3E-25 | 15 | 69 | IPR009057 | Homeodomain-like |
| CDD | cd00167 | 6.96E-10 | 16 | 62 | No hit | No description |
| PROSITE profile | PS51294 | 21.666 | 63 | 117 | IPR017930 | Myb domain |
| SMART | SM00717 | 4.8E-11 | 67 | 115 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 1.3E-10 | 69 | 111 | IPR001005 | SANT/Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 2.9E-23 | 70 | 117 | IPR009057 | Homeodomain-like |
| CDD | cd00167 | 6.42E-8 | 71 | 113 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009751 | Biological Process | response to salicylic acid | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 354 aa Download sequence Send to blast |
MGRAPCCDKA NVKKGPWSPE EDAKLKAYIE QHGTGGNWIA LPQKIGLKRC GKSCRLRWLN 60 YLRPNIKHGG FSEEEDNIIC SLYISIGSRW SIIAAQLPGR TDNDIKNYWN TRLKKKLLGK 120 HRKEIQSRRN NKQISQEMKK GSASRDNYCN NYNNNPYWSA ETVPNNVLPP IPYSNEEPRF 180 NDHTSIRKLL IKLGGRFSSQ DDDDDDQLPI NPHYNIFPAA PEMFSSAPLI DNCLTAAPGP 240 EMECNYKVEG GVDNVAMGST GGGGGESSNF AYELEQMIYN NPQKFEGLEF LCGDIFLMNN 300 YNNNYNRGDF GGGIMSSGWS EMMSSMVLPS SVVSNNSNYE DSGDDHLMRY NAGP |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1a5j_A | 1e-24 | 14 | 117 | 7 | 108 | B-MYB |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription activator (By similarity). Positively regulates axillary meristems (AMs) formation and development, especially during inflorescence. {ECO:0000250, ECO:0000269|PubMed:16461581}. | |||||
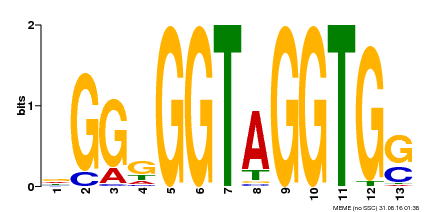
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00089 | SELEX | Transfer from AT3G49690 | Download |
| |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_023878050.1 | 1e-126 | transcription factor RAX3-like isoform X2 | ||||
| Refseq | XP_023884860.1 | 1e-126 | transcription factor RAX3-like isoform X2 | ||||
| Swissprot | Q9M2Y9 | 2e-84 | RAX3_ARATH; Transcription factor RAX3 | ||||
| TrEMBL | J7FAY5 | 1e-124 | J7FAY5_QUESU; R2R3-MYB transcription factor MYB1.1 | ||||
| STRING | cassava4.1_011054m | 1e-123 | (Manihot esculenta) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Asterids | OGEA12 | 24 | 2154 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



