 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | cra_locus_19261_iso_1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Gentianales; Apocynaceae; Rauvolfioideae; Vinceae; Catharanthinae; Catharanthus
|
||||||||
| Family | GATA | ||||||||
| Protein Properties | Length: 341aa MW: 36553.5 Da PI: 6.0548 | ||||||||
| Description | GATA family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | GATA | 54 | 2.2e-17 | 224 | 260 | 1 | 35 |
GATA 1 CsnCgttk..TplWRrgpdgnktLCnaCGlyyrkkgl 35
C++Cgt++ Tp++Rrgp g++tLCnaCGl+++ kg+
cra_locus_19261_iso_1_len_1649_ver_3 224 CTHCGTSSksTPMMRRGPAGPRTLCNACGLFWANKGT 260
**********************************997 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51320 | 13.681 | 91 | 126 | IPR010399 | Tify domain |
| SMART | SM00979 | 3.0E-10 | 91 | 126 | IPR010399 | Tify domain |
| Pfam | PF06200 | 1.8E-11 | 95 | 125 | IPR010399 | Tify domain |
| Pfam | PF06203 | 5.0E-15 | 158 | 199 | IPR010402 | CCT domain |
| PROSITE profile | PS51017 | 12.58 | 158 | 200 | IPR010402 | CCT domain |
| SMART | SM00401 | 8.3E-15 | 218 | 271 | IPR000679 | Zinc finger, GATA-type |
| PROSITE profile | PS50114 | 10.37 | 218 | 275 | IPR000679 | Zinc finger, GATA-type |
| SuperFamily | SSF57716 | 2.14E-12 | 220 | 267 | No hit | No description |
| Gene3D | G3DSA:3.30.50.10 | 1.9E-16 | 223 | 265 | IPR013088 | Zinc finger, NHR/GATA-type |
| CDD | cd00202 | 6.15E-16 | 224 | 270 | No hit | No description |
| PROSITE pattern | PS00344 | 0 | 224 | 251 | IPR000679 | Zinc finger, GATA-type |
| Pfam | PF00320 | 3.9E-15 | 224 | 260 | IPR000679 | Zinc finger, GATA-type |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0007623 | Biological Process | circadian rhythm | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0005515 | Molecular Function | protein binding | ||||
| GO:0008270 | Molecular Function | zinc ion binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| GO:0044212 | Molecular Function | transcription regulatory region DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 341 aa Download sequence Send to blast |
MYAQAQRPVT APSRGQGQVH VKYSPVGDGE DSNAAVCGAA GGESIENAQI VDFDEGSTIG 60 GLDGVEVVPH ESMYNGGAAA VCSDGMALAQ TGDVSNQLTL SFRGQVYVFD SVSTEKVQAV 120 LLLLGGCEYT PGTQGVEGSY QNPRGFTEYV GRCSDPKRAE SLNRFRQKRK ERCFEKKIRY 180 NVRQEVALRM QRKKGQFAAR NSEESMVSDV AEESQNDNQS EILCTHCGTS SKSTPMMRRG 240 PAGPRTLCNA CGLFWANKGT LRDLSKKLQP AAEEDEGDSD SDYGVPIGTS ASISHPNTAF 300 SAGKTSALAA EQXLKTVSPL ACSLITPVTC GMFSTNCRHV W |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcriptional activator that specifically binds 5'-GATA-3' or 5'-GAT-3' motifs within gene promoters. {ECO:0000250, ECO:0000269|PubMed:14966217}. | |||||
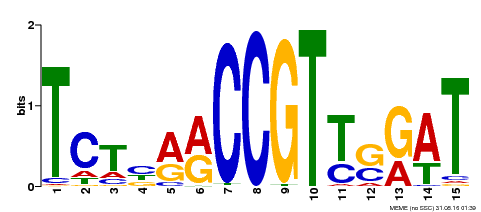
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00451 | DAP | Transfer from AT4G24470 | Download |
| |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_027148698.1 | 1e-133 | GATA transcription factor 25-like | ||||
| Swissprot | Q9LRH6 | 3e-74 | GAT25_ARATH; GATA transcription factor 25 | ||||
| TrEMBL | A0A068TXX6 | 1e-129 | A0A068TXX6_COFCA; Uncharacterized protein | ||||
| STRING | XP_010099559.1 | 6e-91 | (Morus notabilis) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Asterids | OGEA14030 | 10 | 12 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



