 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | cra_locus_22725_iso_1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Gentianales; Apocynaceae; Rauvolfioideae; Vinceae; Catharanthinae; Catharanthus
|
||||||||
| Family | WRKY | ||||||||
| Protein Properties | Length: 618aa MW: 67500.9 Da PI: 6.5439 | ||||||||
| Description | WRKY family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | WRKY | 96.9 | 1.4e-30 | 341 | 399 | 2 | 59 |
--SS-EEEEEEE--TT-SS-EEEEEE-ST.T---EEEEEE-SSSTTEEEEEEES--SS- CS
WRKY 2 dDgynWrKYGqKevkgsefprsYYrCtsa.gCpvkkkversaedpkvveitYegeHnhe 59
Dg++WrKYGqK+ kg+++pr+YYrCt+a gCpv+k+v+r+a+d +++++tYeg+Hnh+
cra_locus_22725_iso_1_len_2006_ver_3 341 TDGCQWRKYGQKMAKGNPCPRAYYRCTMAaGCPVRKQVQRCADDRTILITTYEGNHNHP 399
7***************************99****************************7 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:2.20.25.80 | 1.4E-33 | 325 | 401 | IPR003657 | WRKY domain |
| SuperFamily | SSF118290 | 9.29E-28 | 333 | 401 | IPR003657 | WRKY domain |
| PROSITE profile | PS50811 | 29.792 | 335 | 401 | IPR003657 | WRKY domain |
| SMART | SM00774 | 6.9E-37 | 340 | 400 | IPR003657 | WRKY domain |
| Pfam | PF03106 | 8.2E-27 | 342 | 399 | IPR003657 | WRKY domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0010200 | Biological Process | response to chitin | ||||
| GO:0016036 | Biological Process | cellular response to phosphate starvation | ||||
| GO:0045892 | Biological Process | negative regulation of transcription, DNA-templated | ||||
| GO:0080169 | Biological Process | cellular response to boron-containing substance deprivation | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| GO:0044212 | Molecular Function | transcription regulatory region DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 618 aa Download sequence Send to blast |
MAKGSGLSFD PDPISSPLLF FHNNNNNLHR QKIVKFKEEE EEKEEGVLSM EMDPSVNNRS 60 SPPTIQFPVN LNCNTTTTSH HLDHQEKKIV IDEMDFFADH KKDIIKDIIG DSKASHSTTD 120 HPTATADDAD IRNRTVSTEL DFNVNTGLHL VTGNTSSDQS IVDDGISPHS DDKRAKSELA 180 VLQAELERMN RENRRLKETL NQVTHSYNTL QMHLMTMMQQ QKLEQNVELD QNDGKNEHRL 240 IVPRQFMDLG LANAAVGTAD TDEASLSSSE GRSGYDGSRS PMNNIEGSGR DDSPEKGSQA 300 WCPNKVPRLG HPSKNVDQAT EATMRKARVS VRARSEAPMI TDGCQWRKYG QKMAKGNPCP 360 RAYYRCTMAA GCPVRKQVQR CADDRTILIT TYEGNHNHPL PPAAIAMAST TSSAARMLLS 420 GSMPSADGLM NSNFLARTLL PCSSSMATIS ASAPFPTVTL DLTQNPNPSQ FQRNIPNQFQ 480 LPFPNSPQNL LSNPAAALLP HIFSQALYNQ SKFSGLQMSH DSDVAANNNQ LGHQTMAATT 540 TGLPPLHGHQ GQPNQQNPLA DTLSALTADP NFTAALAAAI TSLIGNSHQN SAPNNNISQN 600 VNSNNNPSVT TSNNNNSH |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 2ayd_A | 5e-22 | 327 | 403 | 1 | 76 | WRKY transcription factor 1 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor involved in the control of processes related to senescence and pathogen defense (PubMed:12000796). Interacts specifically with the W box (5'-(T)TGAC[CT]-3'), a frequently occurring elicitor-responsive cis-acting element (PubMed:12000796). Activates the transcription of the SIRK gene and represses its own expression and that of the WRKY42 genes (PubMed:12000796). Modulates phosphate homeostasis and Pi translocation by regulating PHO1 expression (PubMed:25733771). {ECO:0000269|PubMed:12000796, ECO:0000269|PubMed:25733771}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00209 | DAP | Transfer from AT1G62300 | Download |
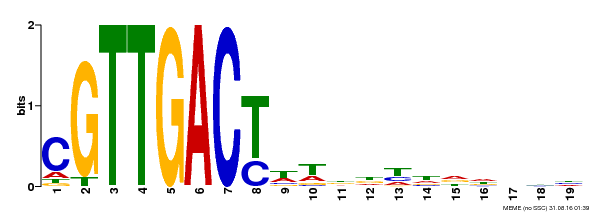
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: By salicylic acid, ethylene, jasmonic acid, pathogens, wounding and strongly during leaf senescence. {ECO:0000269|PubMed:11449049, ECO:0000269|PubMed:11722756}. | |||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_027169937.1 | 0.0 | probable WRKY transcription factor 31 | ||||
| Swissprot | Q9C519 | 1e-143 | WRKY6_ARATH; WRKY transcription factor 6 | ||||
| TrEMBL | A0A068UR90 | 0.0 | A0A068UR90_COFCA; Uncharacterized protein | ||||
| STRING | XP_009614876.1 | 0.0 | (Nicotiana tomentosiformis) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Asterids | OGEA945 | 24 | 92 |



