 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | cra_locus_2287_iso_2 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Gentianales; Apocynaceae; Rauvolfioideae; Vinceae; Catharanthinae; Catharanthus
|
||||||||
| Family | NAC | ||||||||
| Protein Properties | Length: 249aa MW: 27738.4 Da PI: 6.5113 | ||||||||
| Description | NAC family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | NAM | 169.5 | 1.1e-52 | 28 | 155 | 1 | 128 |
NAM 1 lppGfrFhPtdeelvveyLkkkvegkkleleevikevdiykvePwdLp..kkvkaeekewyfFskrdkkyatgk 72
l+pGfrFhPtdeelv +yL++k+ gk++++ ++++e+d+yk+ePw+L +++k+++ ewyfFs+ d+ky +g+
cra_locus_2287_iso_2_len_1244_ver_3 28 LAPGFRFHPTDEELVRYYLRRKACGKPFRF-QAVSEIDVYKSEPWELAcfSSLKTRDLEWYFFSPVDRKYGNGS 100
579*************************99.88**************85347777888**************** PP
NAM 73 rknratksgyWkatgkdkevlskkgelvglkktLvfykgrapkgektdWvmheyrl 128
r nrat +gyWkatgkd++v + k++++g+kktLvf++grap+g++t+Wvmheyrl
cra_locus_2287_iso_2_len_1244_ver_3 101 RLNRATGKGYWKATGKDRPVRH-KNQTIGMKKTLVFHSGRAPDGKRTNWVMHEYRL 155
**********************.9999***************************98 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF101941 | 1.26E-62 | 24 | 178 | IPR003441 | NAC domain |
| PROSITE profile | PS51005 | 57.73 | 28 | 178 | IPR003441 | NAC domain |
| Pfam | PF02365 | 8.8E-29 | 30 | 155 | IPR003441 | NAC domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009555 | Biological Process | pollen development | ||||
| GO:0010228 | Biological Process | vegetative to reproductive phase transition of meristem | ||||
| GO:0005730 | Cellular Component | nucleolus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 249 aa Download sequence Send to blast |
MGQELVAVTP ATPVGGGISG SAPPATSLAP GFRFHPTDEE LVRYYLRRKA CGKPFRFQAV 60 SEIDVYKSEP WELACFSSLK TRDLEWYFFS PVDRKYGNGS RLNRATGKGY WKATGKDRPV 120 RHKNQTIGMK KTLVFHSGRA PDGKRTNWVM HEYRLVDEEL EKAGVPQDTF VLCRIFQKSG 180 LGPPNGDRYA PFIEEEWDDD AALMVVPGGE AEDDMANGDE ARVGCNDLDQ VCFPIGYLLK 240 FPCTAVACS |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 3swm_A | 4e-53 | 17 | 179 | 8 | 169 | NAC domain-containing protein 19 |
| 3swm_B | 4e-53 | 17 | 179 | 8 | 169 | NAC domain-containing protein 19 |
| 3swm_C | 4e-53 | 17 | 179 | 8 | 169 | NAC domain-containing protein 19 |
| 3swm_D | 4e-53 | 17 | 179 | 8 | 169 | NAC domain-containing protein 19 |
| 3swp_A | 4e-53 | 17 | 179 | 8 | 169 | NAC domain-containing protein 19 |
| 3swp_B | 4e-53 | 17 | 179 | 8 | 169 | NAC domain-containing protein 19 |
| 3swp_C | 4e-53 | 17 | 179 | 8 | 169 | NAC domain-containing protein 19 |
| 3swp_D | 4e-53 | 17 | 179 | 8 | 169 | NAC domain-containing protein 19 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcriptional repressor that binds to the motif 5'-(C/T)A(C/A)G-3' in the promoter of target genes (PubMed:25578968). Binds also to the 5'-CTTGNNNNNCAAG-3' consensus sequence in chromatin (PubMed:26617990). Can bind to the mitochondrial dysfunction motif (MDM) present in the upstream regions of mitochondrial dysfunction stimulon (MDS) genes involved in mitochondrial retrograde regulation (MRR) (PubMed:24045019). Together with NAC050 and JMJ14, regulates gene expression and flowering time by associating with the histone demethylase JMJ14, probably by the promotion of RNA-mediated gene silencing (PubMed:25578968, PubMed:26617990). Regulates siRNA-dependent post-transcriptional gene silencing (PTGS) through SGS3 expression modulation (PubMed:28207953). Required during pollen development (PubMed:19237690). {ECO:0000269|PubMed:19237690, ECO:0000269|PubMed:24045019, ECO:0000269|PubMed:25578968, ECO:0000269|PubMed:26617990, ECO:0000269|PubMed:28207953}. | |||||
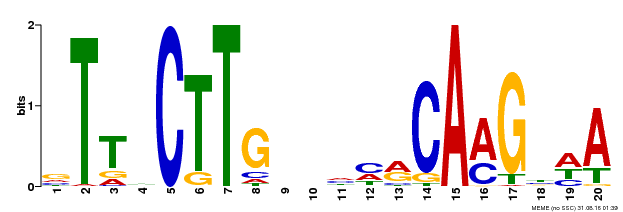
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00612 | ChIP-seq | Transfer from AT3G10490 | Download |
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Induced by type III effector proteins (TTEs) secreted by the pathogenic bacteria P.syringae pv. tomato DC3000 during basal defense. {ECO:0000269|PubMed:16553893}. | |||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_027104903.1 | 1e-147 | NAC domain containing protein 50-like | ||||
| Swissprot | Q9SQY0 | 6e-98 | NAC52_ARATH; NAC domain containing protein 52 | ||||
| TrEMBL | A0A068TWS9 | 1e-146 | A0A068TWS9_COFCA; Uncharacterized protein | ||||
| STRING | XP_009619467.1 | 1e-142 | (Nicotiana tomentosiformis) | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



