 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | cra_locus_23742_iso_1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Gentianales; Apocynaceae; Rauvolfioideae; Vinceae; Catharanthinae; Catharanthus
|
||||||||
| Family | WRKY | ||||||||
| Protein Properties | Length: 359aa MW: 38500.3 Da PI: 4.4809 | ||||||||
| Description | WRKY family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | WRKY | 94.3 | 8.8e-30 | 139 | 196 | 3 | 59 |
-SS-EEEEEEE--TT-SS-EEEEEE-ST.T---EEEEEE-SSSTTEEEEEEES--SS- CS
WRKY 3 DgynWrKYGqKevkgsefprsYYrCtsa.gCpvkkkversaedpkvveitYegeHnhe 59
D ++WrKYGqK++kgs++pr+YYrC+++ gC+++k+ver+ +dp+++++ Y+geH+h+
cra_locus_23742_iso_1_len_1196_ver_3 139 DSWAWRKYGQKPIKGSPYPRNYYRCSTSkGCSARKQVERCPTDPNIFVVSYSGEHTHP 196
99***********************9988****************************8 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:2.20.25.80 | 6.3E-31 | 123 | 198 | IPR003657 | WRKY domain |
| SuperFamily | SSF118290 | 8.89E-27 | 130 | 198 | IPR003657 | WRKY domain |
| PROSITE profile | PS50811 | 32.569 | 132 | 198 | IPR003657 | WRKY domain |
| SMART | SM00774 | 4.0E-35 | 137 | 197 | IPR003657 | WRKY domain |
| Pfam | PF03106 | 1.7E-25 | 139 | 196 | IPR003657 | WRKY domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0007263 | Biological Process | nitric oxide mediated signal transduction | ||||
| GO:0009739 | Biological Process | response to gibberellin | ||||
| GO:0042742 | Biological Process | defense response to bacterium | ||||
| GO:0045892 | Biological Process | negative regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| GO:0044212 | Molecular Function | transcription regulatory region DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 359 aa Download sequence Send to blast |
XAAASSSSSC SINPVAVNDF HVPEVIDLDD KDIIDESFQG LEEIYKELFS PNFSGSVPGE 60 FQLDQRKIQA PPDHEQQVQP AAAGDQLVVQ GQNQHLHIRH GIVCSSSPPQ PAAAGRPRRR 120 KNQQTKTVHQ MTQEELSGDS WAWRKYGQKP IKGSPYPRNY YRCSTSKGCS ARKQVERCPT 180 DPNIFVVSYS GEHTHPRPTH RSSLAGSTRS KLSPSAKQTS TSAATTSINT AAQITSSPPA 240 PATISPTTPI IGGESEGEEN IENDNAEISG NVIEEEEEEE EDQEGNDDEE DILIPNNFIT 300 DDILKGFQEL TGGGGATNNN GSDSSGTFAD KSSCSLLSPP WTPTSSTAGG GCXLAEPNG |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 5w3x_B | 5e-20 | 138 | 199 | 17 | 80 | Disease resistance protein RRS1 |
| 5w3x_D | 5e-20 | 138 | 199 | 17 | 80 | Disease resistance protein RRS1 |
| Search in ModeBase | ||||||
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 114 | 122 | GRPRRRKNQ |
| 2 | 116 | 120 | PRRRK |
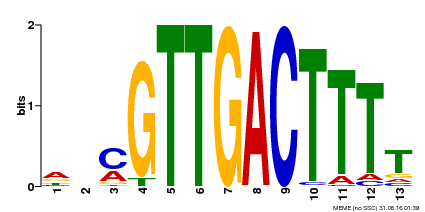
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00561 | DAP | Transfer from AT5G52830 | Download |
| |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| TrEMBL | A0A2G9HFK9 | 4e-80 | A0A2G9HFK9_9LAMI; Uncharacterized protein | ||||
| STRING | EOY20308 | 2e-61 | (Theobroma cacao) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Asterids | OGEA13860 | 10 | 14 |



