 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | cra_locus_24682_iso_2 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Gentianales; Apocynaceae; Rauvolfioideae; Vinceae; Catharanthinae; Catharanthus
|
||||||||
| Family | NAC | ||||||||
| Protein Properties | Length: 335aa MW: 38237.4 Da PI: 4.8827 | ||||||||
| Description | NAC family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | NAM | 173.6 | 5.9e-54 | 6 | 133 | 1 | 128 |
NAM 1 lppGfrFhPtdeelvveyLkkkvegkkleleevikevdiykvePwdLp.k.kvkaeekewyfFskrdkkyatg 71
lppGfrFhPtdeelv+ yLk+k++g ++el evi+ +d+yk++Pw+Lp k + +++kew+fF++rdkky++g
cra_locus_24682_iso_2_len_1443_ver_3 6 LPPGFRFHPTDEELVDXYLKRKTDGLEIEL-EVIPVIDLYKFDPWELPeKsFLPKRDKEWFFFCPRDKKYPNG 77
79****************************.99**************95345566899*************** PP
NAM 72 krknratksgyWkatgkdkevlskkgelvglkktLvfykgrapkgektdWvmheyrl 128
+r+nrat+sgyWkatgkd++v++ +++l+g +ktLvfy+grap g +tdWvmheyrl
cra_locus_24682_iso_2_len_1443_ver_3 78 SRTNRATRSGYWKATGKDRKVVC-QSALIGFRKTLVFYRGRAPLGDRTDWVMHEYRL 133
***********************.999****************************98 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF101941 | 9.29E-62 | 4 | 156 | IPR003441 | NAC domain |
| PROSITE profile | PS51005 | 58.018 | 6 | 156 | IPR003441 | NAC domain |
| Pfam | PF02365 | 5.2E-29 | 7 | 133 | IPR003441 | NAC domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0008283 | Biological Process | cell proliferation | ||||
| GO:0071365 | Biological Process | cellular response to auxin stimulus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 335 aa Download sequence Send to blast |
MGGASLPPGF RFHPTDEELV DXYLKRKTDG LEIELEVIPV IDLYKFDPWE LPEKSFLPKR 60 DKEWFFFCPR DKKYPNGSRT NRATRSGYWK ATGKDRKVVC QSALIGFRKT LVFYRGRAPL 120 GDRTDWVMHE YRLCDDVSQG IPVFQGPFAL CRVIKKNEQK MRDVQGEATF KPVGSSSQTA 180 NVASVEPIVI TDDNPIQASY ICNESNHSTP MTSPYQPTTM GDYDQPSMGT NNPPSLFVSP 240 DMILDSSKDY AQPRGLHPEY GFQNSMWQSS YDQFEISPTS SNSNLQEEAE PSDDFSRYGC 300 MSPYSVYGSY MGYYGNDMLY EGYDQTNSLR NQQPF |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1ut4_A | 3e-53 | 5 | 157 | 16 | 166 | NO APICAL MERISTEM PROTEIN |
| 1ut4_B | 3e-53 | 5 | 157 | 16 | 166 | NO APICAL MERISTEM PROTEIN |
| 1ut7_A | 3e-53 | 5 | 157 | 16 | 166 | NO APICAL MERISTEM PROTEIN |
| 1ut7_B | 3e-53 | 5 | 157 | 16 | 166 | NO APICAL MERISTEM PROTEIN |
| 4dul_A | 3e-53 | 5 | 157 | 16 | 166 | NAC domain-containing protein 19 |
| 4dul_B | 3e-53 | 5 | 157 | 16 | 166 | NAC domain-containing protein 19 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor involved in tissue reunion of wounded inflorescence stems. Required for the division of pith cells in the reunion process, which is dependent on polar-transported auxin and the wound-inducible hormones ethylene and jasmonate (PubMed:21911380). Binds to the promoters of XTH19 and XTH20 to induce their expression via auxin signaling. XTH19 and XTH20 are involved in cell proliferation in the tissue reunion process of incised stems (PubMed:25182467). Involved in hypocotyl graft union formation. Required for the auxin- mediated promotion of vascular tissue proliferation during hypocotyl graft attachment (PubMed:27986917). {ECO:0000269|PubMed:21911380, ECO:0000269|PubMed:25182467, ECO:0000269|PubMed:27986917}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00439 | DAP | Transfer from AT4G17980 | Download |
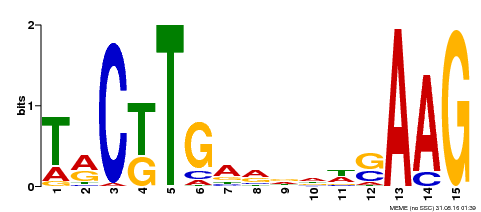
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Induced by wounding in the flowering stem. {ECO:0000269|PubMed:21911380}. | |||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_027080423.1 | 1e-176 | NAC domain-containing protein 71-like | ||||
| Refseq | XP_027183320.1 | 1e-176 | NAC domain-containing protein 71-like | ||||
| Swissprot | O49697 | 3e-94 | NAC71_ARATH; NAC domain-containing protein 71 | ||||
| TrEMBL | A0A068UZB9 | 1e-175 | A0A068UZB9_COFCA; Uncharacterized protein | ||||
| STRING | Solyc08g077110.2.1 | 1e-148 | (Solanum lycopersicum) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Asterids | OGEA6256 | 16 | 35 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



