 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | cra_locus_25985_iso_2 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Gentianales; Apocynaceae; Rauvolfioideae; Vinceae; Catharanthinae; Catharanthus
|
||||||||
| Family | C2H2 | ||||||||
| Protein Properties | Length: 441aa MW: 48043.6 Da PI: 9.3295 | ||||||||
| Description | C2H2 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | zf-C2H2 | 16.6 | 2.1e-05 | 6 | 28 | 1 | 23 |
EEETTTTEEESSHHHHHHHHHHT CS
zf-C2H2 1 ykCpdCgksFsrksnLkrHirtH 23
+ C+ C+k F r nL+ H r H
cra_locus_25985_iso_2_len_1427_ver_3 6 FICEVCNKGFQREQNLQLHRRGH 28
89*******************88 PP
| |||||||
| 2 | zf-C2H2 | 13 | 0.00031 | 82 | 104 | 1 | 23 |
EEETTTTEEESSHHHHHHHHHHT CS
zf-C2H2 1 ykCpdCgksFsrksnLkrHirtH 23
+kC +C+k + +s+ k H +t+
cra_locus_25985_iso_2_len_1427_ver_3 82 FKCDKCSKKYAVQSDWKAHSKTC 104
89*****************9998 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF57667 | 4.64E-8 | 5 | 28 | No hit | No description |
| Gene3D | G3DSA:3.30.160.60 | 4.7E-6 | 5 | 28 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| PROSITE profile | PS50157 | 10.99 | 6 | 28 | IPR007087 | Zinc finger, C2H2 |
| Pfam | PF12171 | 2.0E-5 | 6 | 28 | IPR022755 | Zinc finger, double-stranded RNA binding |
| SMART | SM00355 | 0.0052 | 6 | 28 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE pattern | PS00028 | 0 | 8 | 28 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 130 | 47 | 77 | IPR015880 | Zinc finger, C2H2-like |
| Gene3D | G3DSA:3.30.160.60 | 2.2E-5 | 70 | 103 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| SuperFamily | SSF57667 | 4.64E-8 | 77 | 102 | No hit | No description |
| SMART | SM00355 | 99 | 82 | 102 | IPR015880 | Zinc finger, C2H2-like |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0003676 | Molecular Function | nucleic acid binding | ||||
| GO:0046872 | Molecular Function | metal ion binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 441 aa Download sequence Send to blast |
MATNRFICEV CNKGFQREQN LQLHRRGHNL PWKLRQKTTK EVKRKVYLCP EPTCVHHDPS 60 RALGDLTGIK KHYSRKHGEK KFKCDKCSKK YAVQSDWKAH SKTCGTREYR CDCGXPFLRR 120 DSFITHRAFC DALAQESARN PPSLTGIGSH LFGNTTSNGN NMSLGLSQLS AQISASQDQN 180 QNHSHSASDI LRLGNNSRQF EQHHHHNHNL IGAAGVGSSS TMQGSSAFRT PSQTMPLPSS 240 SFFLPPDHHQ PDDQDYRHHH EHHHQPSLHH HGLMRLSDLH NNVPQSTAPN IFNLSFFPTS 300 MAETTNNVGG GGGLLQLPNP FNNGHNSSGG GGSSNELGSS LFSSSGSLVG GGGDMTSAIP 360 SLYSSSILHH HQSHGDVGHM SATALLQKAA QLGSTTSNSS ASSLLKSFGS TTSNGSVFGT 420 ATDHHHHHQN NVNGGNNNNH R |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 5b3h_C | 3e-30 | 78 | 139 | 3 | 64 | Zinc finger protein JACKDAW |
| 5b3h_F | 3e-30 | 78 | 139 | 3 | 64 | Zinc finger protein JACKDAW |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor acting as a positive regulator of the starch synthase SS4. Controls chloroplast development and starch granule formation (PubMed:22898356). Binds DNA via its zinc fingers (PubMed:24821766). Recognizes and binds to SCL3 promoter sequence 5'-AGACAA-3' to promotes its expression when in complex with RGA (PubMed:24821766). {ECO:0000269|PubMed:22898356, ECO:0000269|PubMed:24821766}. | |||||
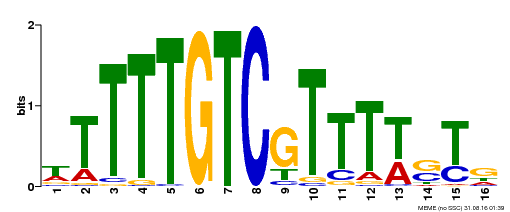
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00255 | DAP | Transfer from AT2G02070 | Download |
| |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_021691809.1 | 1e-142 | protein indeterminate-domain 5, chloroplastic-like | ||||
| Swissprot | Q9ZUL3 | 1e-105 | IDD5_ARATH; Protein indeterminate-domain 5, chloroplastic | ||||
| TrEMBL | A0A2K1ZH36 | 1e-139 | A0A2K1ZH36_POPTR; Uncharacterized protein | ||||
| TrEMBL | B9S7A9 | 1e-139 | B9S7A9_RICCO; Nucleic acid binding protein, putative | ||||
| STRING | POPTR_0008s14180.4 | 1e-139 | (Populus trichocarpa) | ||||
| STRING | XP_002521878.1 | 1e-140 | (Ricinus communis) | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



